Appendix C — Extras
Below is some more in-depth information into specific areas associated with standardizing your data to Darwin Core and uploading it through the IPT.
C.1 Ecological Metadata Language (EML)
The Ecological Metadata Language (EML) is a community developed and maintained metadata standard that is typically associated with ecological-, and earth and environmental data. The purpose of EML is to provide the ecological community with an extensible, flexible, metadata standard used in data analysis and archiving, which will allow automated machine processing, searching and retrieval. EML has been around since 2003, and can be considered a “dialect” or specification to XML to describe tables and other data objects. The XML Schema provides a framework for the metadata, with defined “rules” on how to organize the metadata without any stipulations (another way to put it, XML is the language that defines the rules that govern the EML syntax). The XML Schema defines the structure of some information in a document (e.g. elements and attributes names and relationships), but does not provide any specific details the information included within. An EML document or file (eml.xml) is used to provide detailed description of metadata related to data objects, including tables (and other data objects), their columns, typing etc, and how data tables are linked or grouped. EML is widely used for datasets about ecosystem level observations, and can be used to detail data table information to a high granularity, which allows data users to arrange data tables in any way they need to. EML is particularly useful for wide data tables as table level details are entirely contained within the metadata document, meaning that there is not necessarily a need for external definitions such as a code list. However, if you have a long table arrangement, like the Darwin Core Archive (DwC-A), you can define the allowable values for the column in the metadata as well.
EML is excellent at describing the details of a column of data so that the data values in the tables can be read into analysis systems or an analysis environment using the metadata, or even into a relational database. EML allows for tables to be easily reusable, and read into workflows, translated or reformatted. A drawback to EML is that, compared to the ISO standard, EML is a community standard, adopting a more ‘bottom-up’ approach. This is contrary to the ISO standard, which are internationally agreed upon standards by experts (‘top-down’). However, at the time of EML development in the early 2000’s there was a gap in metadata options to describe ecological data tables, with ISO standards typically being more applicable to geographic data. EML can cover almost anything and is particularly good at tabular data. But at the same time, due to the self-contained nature, there can be little control from outside lists, which means that the description is left to the EML constructor (data provider/manager) and consequently, individual datasets can look quite different from each other, even when they contain similar measurements. As such, it will be important to document best practices and clear mapping of fields between different metadata schemas (e.g. cross-walks between ISO.xml and EML.xml). As of version 2.2, EML can link to external ontologies, and there is capacity for annotation with external terms (e.g. through their URIs). Code lists and external dictionaries can help as they sometimes contain additional information that might not fit into EML (e.g. protocols, or code lists stored in ontologies). Having these external code lists and exporting them as EML snippets could go a long way in reducing that heterogeneity, because the constructors can then select measurements from lists when developing EML documents.
EML is implemented as a series of XML document types (modules) that can be used in an extensible manner to document ecological data. Each EML module is designed to describe one logical part of the total metadata that should be included with any ecological dataset. The architecture of EML was designed to serve the needs of the ecological community, and has benefitted from previous work in other related metadata standards. Using this format can facilitate future growth of the metadata language, and EML supports an active developer community (see e.g. NCEAS EML GitHub). EML adopts much of its syntax from the other metadata standards that have evolved from the expertise of groups in other disciplines. Whenever possible, EML adopted entire trees of information in order to facilitate conversion of EML documents into other metadata languages. The GBIF IPT is a tool used to create a single eml.xml file format inside the DwC-A data package. However, the IPT does not use any of the EML’s built-in table description modules, and perhaps primarily uses one EML module (resource module) for high-level metadata.
However, it is important to know how both OBIS and GBIF use EML, as often a higher granularity of the metadata can be found in the original data tables. An example of this is spatial coverage. The IPT only allows for either a bounding box to be documented (populating North, South, East, and West coordinates), or a single polygon. The EML document however would be able to capture multiple polygons worth of spatial coverage (i.e. a polygon for each transect surveyed). This more detailed information however is often captured in the data (in an OBIS record). Additionally, not all fields that can be populated in an EML document can be translated to the IPT, or harvested by OBIS and GBIF. The GBIF IPT only produces a select number of fields or attributes available in EML.
Important: When reading the EML section in the OBIS manual, you’ll notice that it reads that OBIS uses the GBIF EML profile (version 1.1). However, the current EML version is 2.2.0, as per EcoInformatics. This does not mean that these versions are not compatible, rather, it means that the GBIF IPT currently uses a subset of available EML 2.2.0 fields and attributes, the subset of which they have versioned 1.1.
If you are interested in creating an EML metadata file, it is possible to upload those into the IPT. There are R packages that can help in developing an EML.xml file. These packages are e.g. EML, emld or EMLassemblyline.
C.2 Example using GitHub to resolve errors
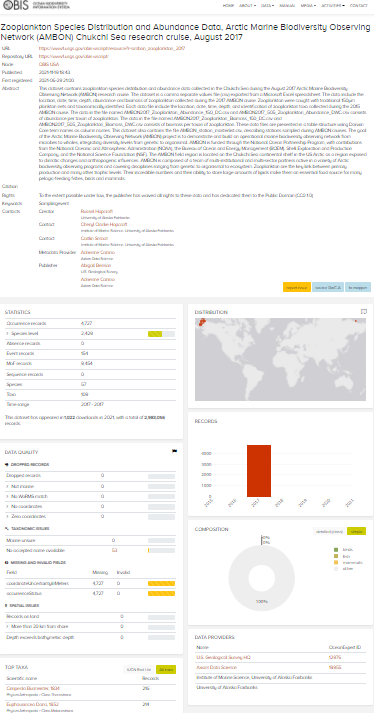
- Dataset sent to OBIS-USA via email.
- OBIS-USA uploaded to IPT.
- Once the data were uploaded, the IPT identified there was an issue with the
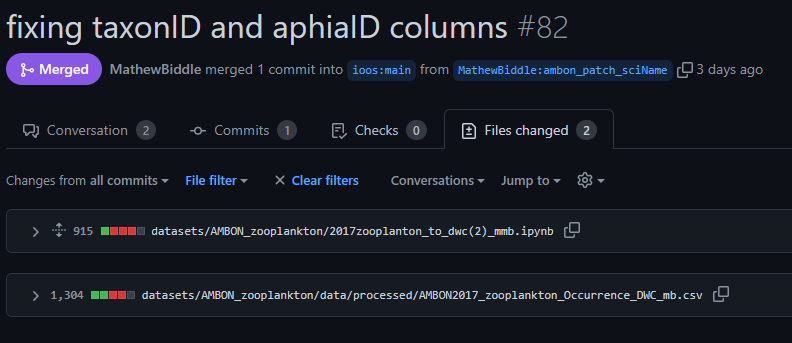
occurrenceIDfield. The issue was then presented and discussed in a GitHub ticket: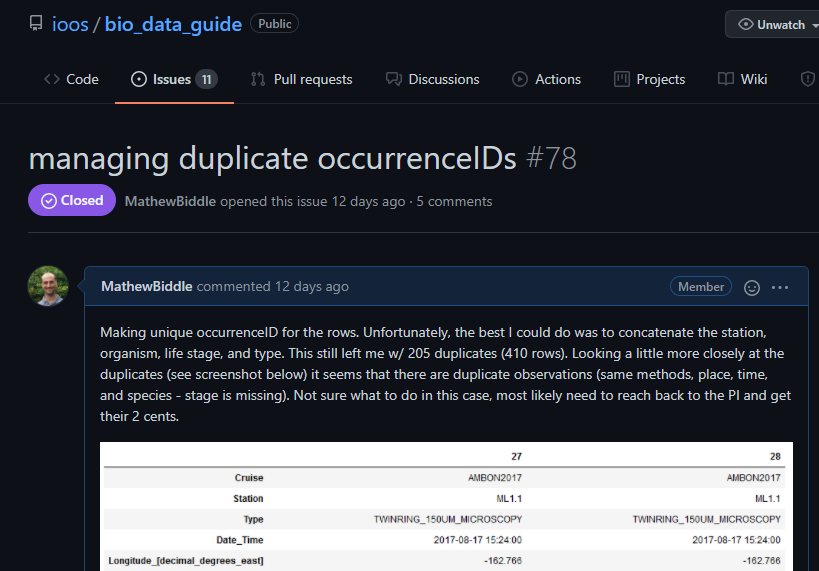
- The data manager uploaded the raw data and code to GitHub through the pull request below. This included a fix for the
occurrenceIDissue.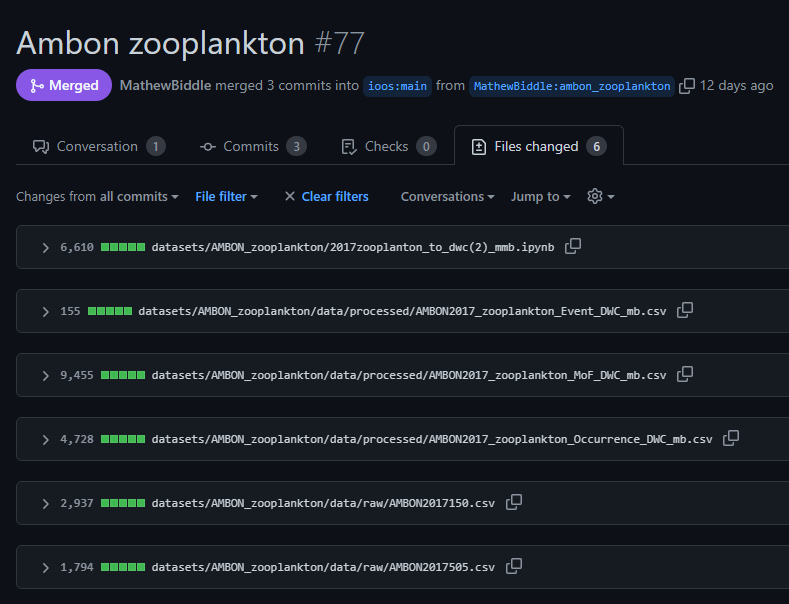
- The OBIS node manager was notified of the availability of a revised dataset by pointing directly to the appropriate commit in GitHub:
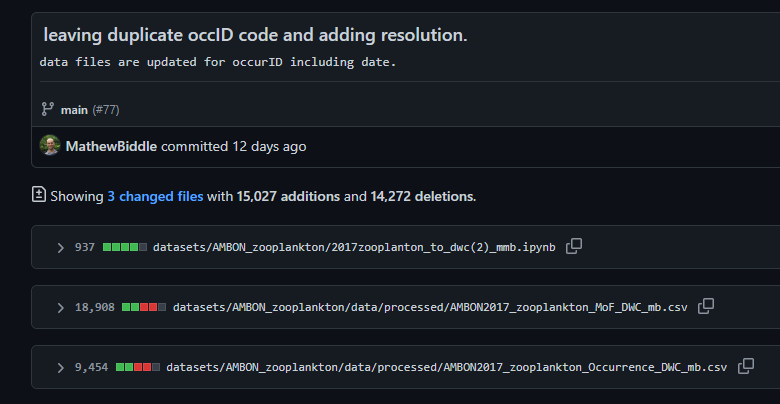
- The OBIS node manager downloaded the data from the commit above and uploaded them to the IPT.
- The IPT returned a summary of the dataset including that 434 records had invalid
scientificNameIDrecords in the occurrence file. - After some data sleuthing, the data manager noticed that the code accidentally removed trailing zeros from
scientificNameIDthat ended in0: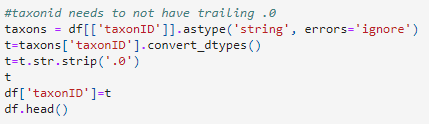
- So, the data manager updated the code to resolve the issue and generate a new occurrence file.
- Here is fixing the
scientificNameIDgeneration: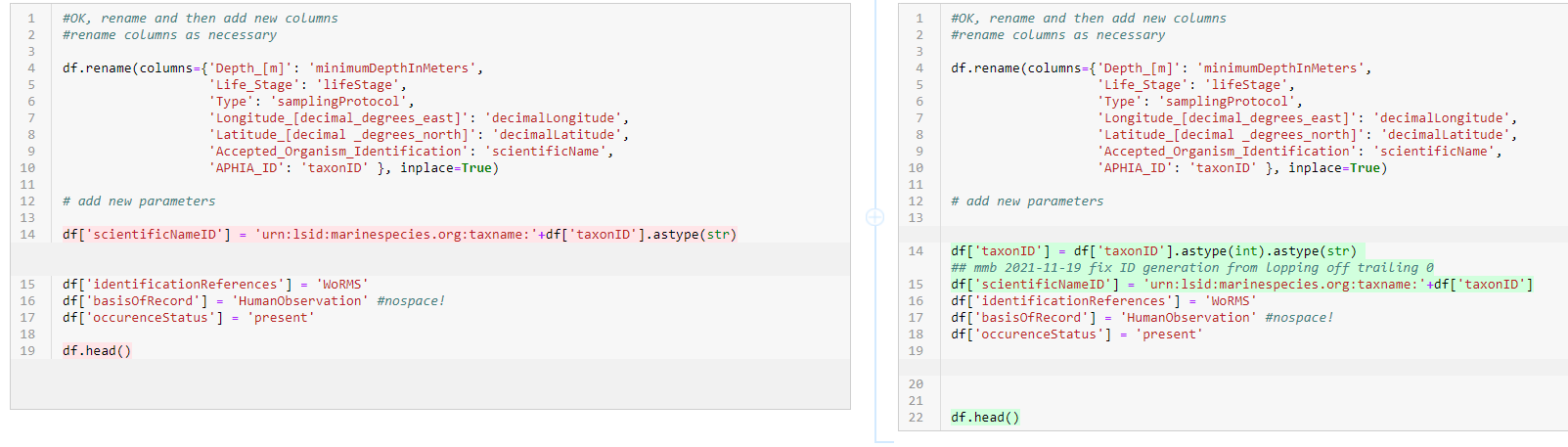
- Here is removing the problematic code:
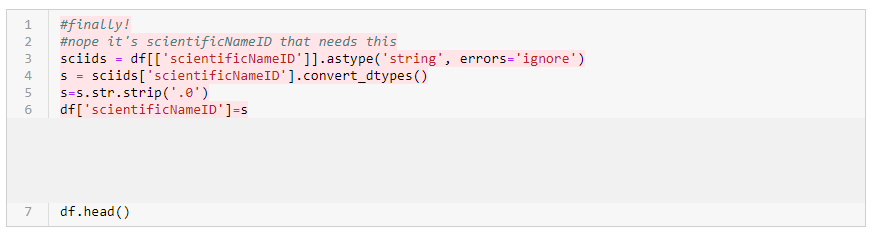

- Here is fixing the
- The revised occurrence file was then resubmitted to the OBIS node manager by pointing them at the appropriate commit record:
- The OBIS node manager downloaded the data from the commit above and uploaded them to the IPT.
- The IPT and OBIS landing page now indicated that no more issues with these data are present: