Calling R libraries from Python#
Created: 2017-11-30
In this example we will explore the Coral Reef Evaluation and Monitoring Project (CREMP) data available in the Gulf of Mexico Coastal Ocean Observing System (GCOOS) ERDDAP server.
To access the server we will use the rerddap library and export the data to Python for easier plotting.
The first step is to load the rpy2 extension that will allow us to use the R libraries.
%load_ext rpy2.ipython
The first line below has a %%R to make it an R cell. The code below specify the GCOOS server and fetches the data information for the fk_CREMP_yearly_revisited_DATA_v3_1996 dataset.
For more information on rerddap please see https://docs.ropensci.org/rerddap/.
%%R
library('rerddap')
url <- 'https://gcoos4.tamu.edu/erddap'
data_info <- rerddap::info('fk_CREMP_yearly_revisited_DATA_v3_1996', url=url)
data_info
<ERDDAP info> fk_CREMP_yearly_revisited_DATA_v3_1996
Base URL: https://gcoos4.tamu.edu/erddap
Dataset Type: tabledap
Variables:
acceptedNameAuthorship:
acceptedNameUsage:
acceptedNameUsageID:
averageNumberOfPoints:
Range: 407, 900
Units: count
basisOfRecord:
bottomType:
class:
country:
crs:
datasetID:
datasetName:
depth:
Range: 3.0, 54.0
Units: m
eventDate:
Range: 1996, 1996
Units: unknown
eventID:
family:
firstYear:
Range: 1996, 1996
Units: unknown
genus:
geodeticDatum:
habitat:
habitatID:
kingdom:
language:
latitude:
Range: 24.4517, 25.2953
Units: degrees_north
license:
locality:
longitude:
Range: -81.9195, -80.2087
Units: degrees_east
maximumDepthInMeters:
Range: 3.0, 54.0
Units: m
minimumDepthInMeters:
Range: 3.0, 54.0
Units: m
Observations:
Range: 0, 11839
occurrenceID:
occurrenceStatus:
order:
organismQuantity:
Range: 0.0, 0.9475
Units: 0.01percent
organismQuantityType:
ownerInstitutionCode:
phylum:
recordedBy:
samplingEffort:
samplingProtocol:
scientificName:
scientificNameAuthorship:
scientificNameID:
siteCode:
siteID:
Range: 10, 81
Units: unknown
siteName:
specificEpithet:
stateProvince:
stationNumber:
Range: 101, 814
Units: unknown
subRegionID:
taxonomicStatus:
taxonRank:
time:
Range: 8.283168E8, 8.283168E8
Units: seconds since 1970-01-01T00:00:00Z
transectLengthInMeters:
Range: 19.0, 26.0
Units: m
type:
vernacularName:
waterBody:
By inspecting the information above we can find the variables available in the dataset and use the tabledap function to download them.
Note that the %%R -o rdf will export the rdf variable back to the Python workspace.
%%R -o df
fields <- c(
'depth',
'longitude',
'latitude',
'organismQuantity',
'genus',
'habitat'
)
df <- tabledap(
data_info,
fields=fields,
url=url
)
R[write to console]: info() output passed to x; setting base url to: https://gcoos4.tamu.edu/erddap
Now we need to export the R DataFrame to a pandas objects and ensure that all numeric types are numbers and not strings.
import pandas as pd
cols = ["longitude", "latitude", "depth", "organismQuantity"]
df[cols] = df[cols].apply(pd.to_numeric)
df.head()
| depth | longitude | latitude | organismQuantity | genus | habitat | |
|---|---|---|---|---|---|---|
| 2 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 3 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 4 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 5 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 6 | 9.0 | -80.3782 | 25.1201 | 0.0 | Acropora | Hard Bottom |
We can navigate to ERDDAP’s info page to find the variables description. Let’s check what is organismQuantity:
The is value of the derived information product, such as the numerical value for biomass. This term does not include units. Mean number of observed fish per species for 5 Minutes
We can see that organismQuantity has a lot of zero values,
let’s remove that first to plot the data positions only where something was found.
# Filter invalid values (-999).
cremp_1996 = df.loc[df["organismQuantity"] >= 0]
cremp_1996["genus"] = cremp_1996["genus"].str.strip()
cremp_1996.head()
| depth | longitude | latitude | organismQuantity | genus | habitat | |
|---|---|---|---|---|---|---|
| 2 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 3 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 4 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 5 | 6.0 | -80.3475 | 25.1736 | 0.0 | Acropora | Hard Bottom |
| 6 | 9.0 | -80.3782 | 25.1201 | 0.0 | Acropora | Hard Bottom |
What is the most common genus of Coral observed?
avg = cremp_1996.drop("habitat", axis=1).groupby("genus").mean()
avg
| depth | longitude | latitude | organismQuantity | |
|---|---|---|---|---|
| genus | ||||
| 23.2375 | -81.046667 | 24.768958 | 0.034553 | |
| Acropora | 23.2375 | -81.046667 | 24.768957 | 0.006003 |
| Agaricia | 23.2375 | -81.046667 | 24.768957 | 0.000005 |
| Cladocora | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Colpophyllia | 23.2375 | -81.046667 | 24.768957 | 0.005079 |
| Dendrogyra | 23.2375 | -81.046667 | 24.768957 | 0.001327 |
| Diadema | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Dichocoenia | 23.2375 | -81.046667 | 24.768957 | 0.000591 |
| Dictyota | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Diploria | 23.2375 | -81.046667 | 24.768957 | 0.001321 |
| Eusmilia | 23.2375 | -81.046667 | 24.768957 | 0.000042 |
| Favia | 23.2375 | -81.046667 | 24.768957 | 0.000031 |
| Gorgonia | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Halimeda | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Helioseris | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Isophyllia | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Lobophora | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Lyngbia | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Madracis | 23.2375 | -81.046667 | 24.768957 | 0.000018 |
| Manicina | 23.2375 | -81.046667 | 24.768957 | 0.000021 |
| Meandrina | 23.2375 | -81.046667 | 24.768957 | 0.000421 |
| Millepora | 23.2375 | -81.046667 | 24.768957 | 0.006130 |
| Montastraea | 23.2375 | -81.046667 | 24.768957 | 0.013394 |
| Mussa | 23.2375 | -81.046667 | 24.768957 | 0.000051 |
| Mycetophyllia | 23.2375 | -81.046667 | 24.768957 | 0.000133 |
| Oculina | 23.2375 | -81.046667 | 24.768957 | 0.000065 |
| Orbicella | 23.2375 | -81.046667 | 24.768957 | 0.034583 |
| Palythoa | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Phyllangia | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Porites | 23.2375 | -81.046667 | 24.768957 | 0.003351 |
| Pseudodiploria | 23.2375 | -81.046667 | 24.768957 | 0.001383 |
| Scolymia | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
| Siderastrea | 23.2375 | -81.046667 | 24.768957 | 0.004472 |
| Solenastrea | 23.2375 | -81.046667 | 24.768957 | 0.000051 |
| Stephanocoenia | 23.2375 | -81.046667 | 24.768957 | 0.000271 |
| Undaria | 23.2375 | -81.046667 | 24.768957 | 0.001139 |
| Xestospongia | 23.2375 | -81.046667 | 24.768957 | 0.000000 |
rerddap’s info request does not have enough metadata about the variables to explain the blank, and most abundant, genus. Checking the sever did not help figure that out. We’ll remove that for now to deal with only those that are identified.
cremp_1996 = cremp_1996.loc[cremp_1996["genus"] != ""]
There are also many genus with zero biomass count. In this example we’ll choose to do a biased analysis of occurrence and eliminate those where nothing was observed.
# Filter zero values (nothing was observed).
cremp_1996 = cremp_1996.loc[cremp_1996["organismQuantity"] > 0]
Now we can check the quantificationValue average by genus.
%matplotlib inline
avg = (
cremp_1996.drop("habitat", axis=1).groupby("genus").mean()
) # re-compute the "biased" average.
ax = avg["organismQuantity"].plot(kind="bar")
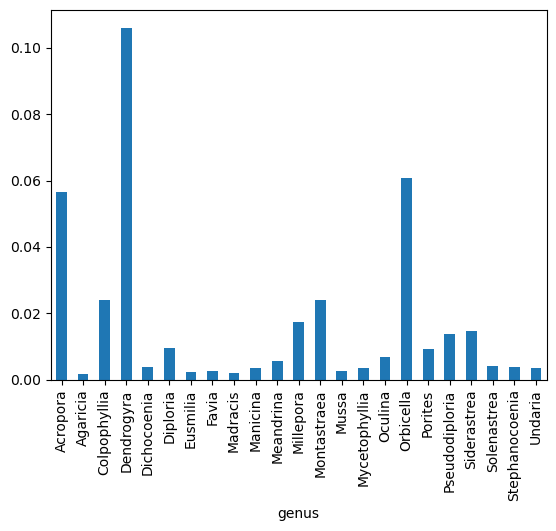
and habitat.
ax = (
cremp_1996.drop("genus", axis=1)
.groupby("habitat")
.mean()["organismQuantity"]
.plot(kind="bar")
)
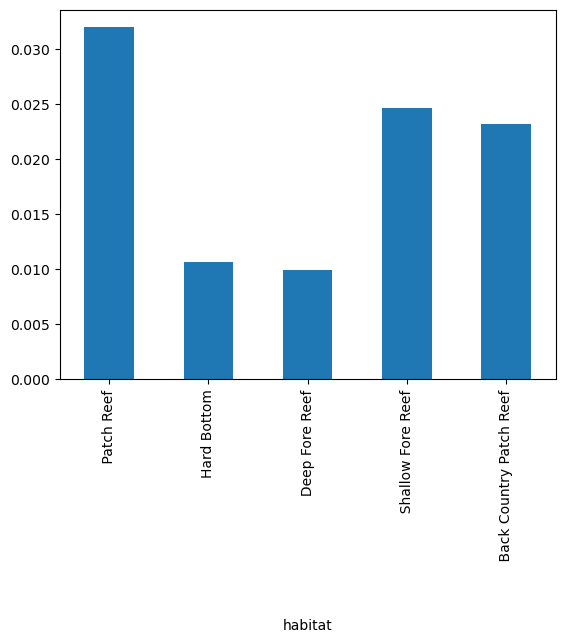
It seems the most of the biomass was found around the Dendrogyra genus in Patch Reef habitats.
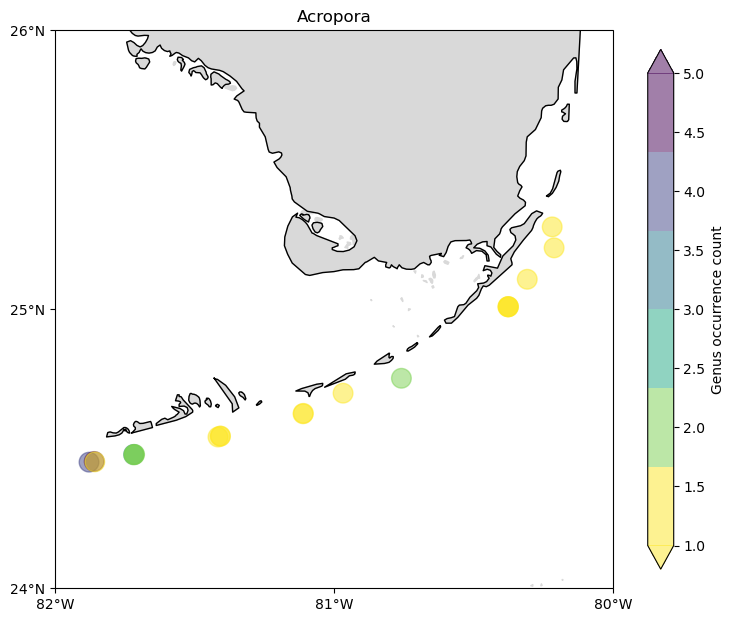
But where are those Coral Reefs? How is the distribution of top three species with more biomass around them?
With a pandas DataFrame it is easy to group the data by location and count the genus occurrence based on it.
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import numpy as np
from cartopy.mpl.ticker import LatitudeFormatter, LongitudeFormatter
def make_plot():
bbox = [-82, -80, 24, 26]
projection = ccrs.PlateCarree()
fig, ax = plt.subplots(figsize=(9, 9), subplot_kw=dict(projection=projection))
ax.set_extent(bbox)
land = cfeature.NaturalEarthFeature(
"physical", "land", "10m", edgecolor="face", facecolor=[0.85] * 3
)
ax.add_feature(land, zorder=0)
ax.coastlines("10m", zorder=1)
ax.set_xticks(np.linspace(bbox[0], bbox[1], 3), crs=projection)
ax.set_yticks(np.linspace(bbox[2], bbox[3], 3), crs=projection)
lon_formatter = LongitudeFormatter(zero_direction_label=True)
lat_formatter = LatitudeFormatter()
ax.xaxis.set_major_formatter(lon_formatter)
ax.yaxis.set_major_formatter(lat_formatter)
return fig, ax
count = (
cremp_1996.loc[cremp_1996["genus"] == "Acropora"]
.groupby(["longitude", "latitude"])
.count()
.reset_index()
)
fig, ax = make_plot()
c = ax.scatter(
count["longitude"],
count["latitude"],
s=200,
c=count["genus"],
alpha=0.5,
cmap=plt.cm.get_cmap("viridis_r", 6),
zorder=3,
)
cbar = fig.colorbar(c, shrink=0.75, extend="both")
cbar.ax.set_ylabel("Genus occurrence count")
ax.set_title("Acropora");
/tmp/ipykernel_9861/710676339.py:16: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap(obj)`` instead.
cmap=plt.cm.get_cmap("viridis_r", 6),
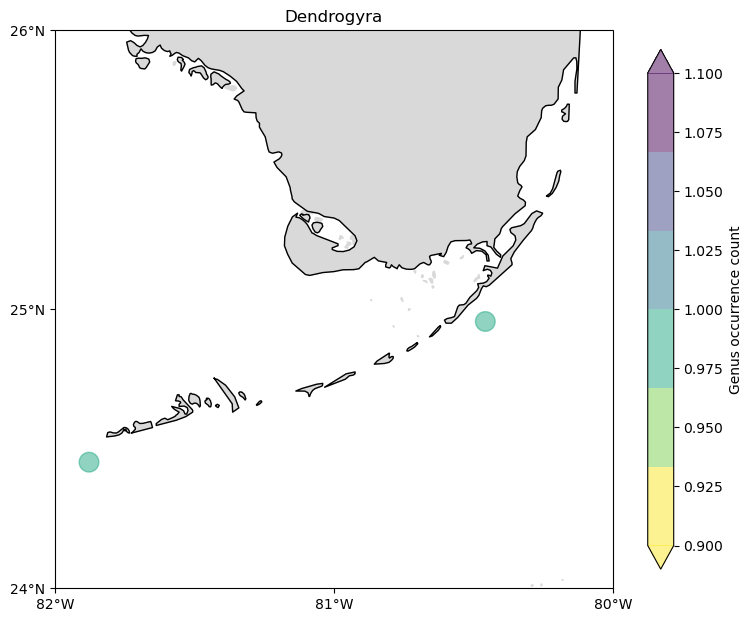
count = (
cremp_1996.loc[cremp_1996["genus"] == "Dendrogyra"]
.groupby(["longitude", "latitude"])
.count()
.reset_index()
)
fig, ax = make_plot()
c = ax.scatter(
count["longitude"],
count["latitude"],
s=200,
c=count["genus"],
alpha=0.5,
cmap=plt.cm.get_cmap("viridis_r", 6),
zorder=3,
)
cbar = fig.colorbar(c, shrink=0.75, extend="both")
cbar.ax.set_ylabel("Genus occurrence count")
ax.set_title("Dendrogyra");
/tmp/ipykernel_9861/1312933967.py:16: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap(obj)`` instead.
cmap=plt.cm.get_cmap("viridis_r", 6),
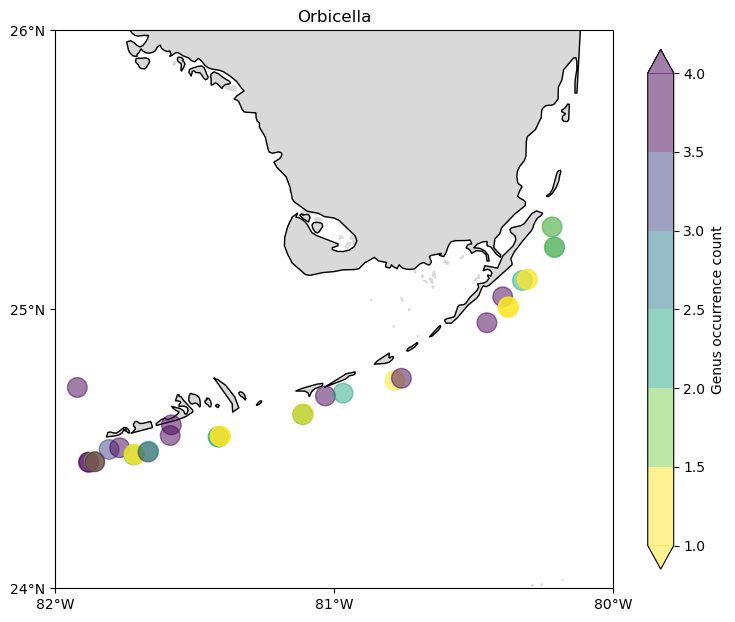
count = (
cremp_1996.loc[cremp_1996["genus"] == "Orbicella"]
.groupby(["longitude", "latitude"])
.count()
.reset_index()
)
fig, ax = make_plot()
c = ax.scatter(
count["longitude"],
count["latitude"],
s=200,
c=count["genus"],
alpha=0.5,
cmap=plt.cm.get_cmap("viridis_r", 6),
zorder=3,
)
cbar = fig.colorbar(c, shrink=0.75, extend="both")
cbar.ax.set_ylabel("Genus occurrence count")
ax.set_title("Orbicella");
/tmp/ipykernel_9861/1632937538.py:16: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap(obj)`` instead.
cmap=plt.cm.get_cmap("viridis_r", 6),
This demonstration showed the power of mixing Python and R to reduce developer time and allow the research to focus on the data and not the programming language.


