Programmatically query the IOOS Data Catalog for a specific observation type#
Created: 2024-09-17
Updated: 2025-03-06
Author: Mathew Biddle
In this notebook we highlight the ability to search the IOOS Data Catalog for a specific subset of observations using the CKAN web accessible Application Programming Interface (API).
For this example, we want to look for observations of oxygen in the water column across the IOOS Catalog. As part of the IOOS Metadata Profile, which the US IOOS community uses to publish datasets, we know that each Regional Association and DAC will be following the Climate and Forecast (CF) Conventions and using CF standard_names to describe their datasets. So, with that assumption, we can search across the IOOS Data catalog for datasets with the CF standard names that contain oxygen and sea_water. Then, we can build a simple map to show the geographical distribution of those datasets.
Build CKAN API query base.#
Uses ckan/ckanapi
from ckanapi import RemoteCKAN
ioos_catalog = RemoteCKAN(
address="https://data.ioos.us",
user_agent="ckanapiioos/1.0 (+https://ioos.us/)",
)
ioos_catalog
<ckanapi.remoteckan.RemoteCKAN at 0x7aa69aba7e00>
What organizations are in the catalog?#
Tell me what organizations are there.
orgs = ioos_catalog.action.organization_list()
print(orgs)
['aoos', 'caricoos', 'cdip', 'cencoos', 'comt', 'gcoos', 'glider-dac', 'glos', 'hf-radar-dac', 'ioos', 'maracoos', 'nanoos', 'neracoos', 'noaa-co-ops', 'noaa-ndbc', 'oceansites', 'pacioos', 'sccoos', 'secoora', 'unidata', 'usgs', 'us-navy']
How many datasets are we searching across?#
Grab all the datasets available and return the count.
datasets = ioos_catalog.action.package_search()
datasets["count"]
36396
Grab the most recent applicable CF standard names#
Collect CF standard names that contain oxygen and sea_water from the CF standard name list.
import pandas as pd
url = "https://cfconventions.org/Data/cf-standard-names/current/src/cf-standard-name-table.xml"
tbl_version = pd.read_xml(url, xpath="./*")["version_number"][0].astype(int)
df = pd.read_xml(url, xpath="entry")
std_names = df.loc[
(df["id"].str.contains("oxygen") & df["id"].str.contains("sea_water"))
]
print(f"CF Standard Name Table: {tbl_version}")
std_names[["id", "description"]]
CF Standard Name Table: 90
| id | description | |
|---|---|---|
| 471 | depth_at_shallowest_local_minimum_in_vertical_... | Depth is the vertical distance below the surfa... |
| 627 | fractional_saturation_of_oxygen_in_sea_water | Fractional saturation is the ratio of some mea... |
| 1368 | mass_concentration_of_oxygen_in_sea_water | Mass concentration means mass per unit volume ... |
| 1737 | mole_concentration_of_dissolved_molecular_oxyg... | Mole concentration means number of moles per u... |
| 1738 | mole_concentration_of_dissolved_molecular_oxyg... | "Mole concentration at saturation" means the m... |
| 1739 | mole_concentration_of_dissolved_molecular_oxyg... | Mole concentration means number of moles per u... |
| 1838 | mole_concentration_of_preformed_dissolved_mole... | "Mole concentration" means the number of moles... |
| 2009 | moles_of_oxygen_per_unit_mass_in_sea_water | moles_of_X_per_unit_mass_inY is also called "m... |
| 3225 | surface_molecular_oxygen_partial_pressure_diff... | The surface called "surface" means the lower b... |
| 3725 | temperature_of_sensor_for_oxygen_in_sea_water | Temperature_of_sensor_for_oxygen_in_sea_water ... |
| 4825 | volume_fraction_of_oxygen_in_sea_water | "Volume fraction" is used in the construction ... |
| 4830 | volume_mixing_ratio_of_oxygen_at_stp_in_sea_water | "ratio_of_X_to_Y" means X/Y. "stp" means stand... |
Search across IOOS Data Catalog using CKAN API#
Search the IOOS Data Catalog for CF standard names that match those above.
import time
from ckanapi import RemoteCKAN
from ckanapi.errors import CKANAPIError
from requests.exceptions import ChunkedEncodingError
from urllib3.exceptions import IncompleteRead
ua = "ckanapiioos/1.0 (+https://ioos.us/)"
ioos_catalog = RemoteCKAN("https://data.ioos.us", user_agent=ua)
ioos_catalog
df_out = pd.DataFrame()
for std_name in std_names["id"]:
print(std_name)
fq = f"+cf_standard_names:{std_name}"
result_count = 0
df_std_name = pd.DataFrame()
while True:
try:
datasets = ioos_catalog.action.package_search(
fq=fq, rows=500, start=result_count
)
except (CKANAPIError, IncompleteRead, ChunkedEncodingError):
continue
num_results = datasets["count"]
print(f"num_results: {num_results}, result_count: {result_count}")
for dataset in datasets["results"]:
df = pd.DataFrame(
{
"title": [dataset["title"]],
"url": [dataset["resources"][0]["url"]],
"org": [dataset["organization"]["title"]],
"std_name": std_name,
}
)
df_std_name = pd.concat([df_std_name, df], ignore_index=True)
result_count = df_std_name.shape[0]
time.sleep(1)
if result_count >= num_results:
print(f"num_results: {num_results}, result_count: {result_count}")
break
df_out = pd.concat([df_out, df_std_name], ignore_index=True)
print(
f"num_results: {num_results}, result_count: {result_count}, total_result_count: {df_out.shape[0]}"
)
df_out.shape
depth_at_shallowest_local_minimum_in_vertical_profile_of_mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
num_results: 0, result_count: 0
num_results: 0, result_count: 0
num_results: 0, result_count: 0, total_result_count: 0
fractional_saturation_of_oxygen_in_sea_water
num_results: 5110, result_count: 0
num_results: 5110, result_count: 500
num_results: 5110, result_count: 1000
num_results: 5110, result_count: 1500
num_results: 5110, result_count: 2000
num_results: 5110, result_count: 2500
num_results: 5110, result_count: 3000
num_results: 5110, result_count: 3500
num_results: 5110, result_count: 4000
num_results: 5110, result_count: 4500
num_results: 5110, result_count: 5000
num_results: 5110, result_count: 5110
num_results: 5110, result_count: 5110, total_result_count: 5110
mass_concentration_of_oxygen_in_sea_water
num_results: 4107, result_count: 0
num_results: 4107, result_count: 500
num_results: 4107, result_count: 1000
num_results: 4107, result_count: 1500
num_results: 4107, result_count: 2000
num_results: 4107, result_count: 2500
num_results: 4107, result_count: 3000
num_results: 4107, result_count: 3500
num_results: 4107, result_count: 4000
num_results: 4107, result_count: 4107
num_results: 4107, result_count: 4107, total_result_count: 9217
mole_concentration_of_dissolved_molecular_oxygen_in_sea_water
num_results: 352, result_count: 0
num_results: 352, result_count: 352
num_results: 352, result_count: 352, total_result_count: 9569
mole_concentration_of_dissolved_molecular_oxygen_in_sea_water_at_saturation
num_results: 0, result_count: 0
num_results: 0, result_count: 0
num_results: 0, result_count: 0, total_result_count: 9569
mole_concentration_of_dissolved_molecular_oxygen_in_sea_water_at_shallowest_local_minimum_in_vertical_profile
num_results: 0, result_count: 0
num_results: 0, result_count: 0
num_results: 0, result_count: 0, total_result_count: 9569
mole_concentration_of_preformed_dissolved_molecular_oxygen_in_sea_water
num_results: 0, result_count: 0
num_results: 0, result_count: 0
num_results: 0, result_count: 0, total_result_count: 9569
moles_of_oxygen_per_unit_mass_in_sea_water
num_results: 796, result_count: 0
num_results: 796, result_count: 500
num_results: 796, result_count: 796
num_results: 796, result_count: 796, total_result_count: 10365
surface_molecular_oxygen_partial_pressure_difference_between_sea_water_and_air
num_results: 0, result_count: 0
num_results: 0, result_count: 0
num_results: 0, result_count: 0, total_result_count: 10365
temperature_of_sensor_for_oxygen_in_sea_water
num_results: 213, result_count: 0
num_results: 213, result_count: 213
num_results: 213, result_count: 213, total_result_count: 10578
volume_fraction_of_oxygen_in_sea_water
num_results: 2183, result_count: 0
num_results: 2183, result_count: 500
num_results: 2183, result_count: 1000
num_results: 2183, result_count: 1500
num_results: 2183, result_count: 2000
num_results: 2183, result_count: 2183
num_results: 2183, result_count: 2183, total_result_count: 12761
volume_mixing_ratio_of_oxygen_at_stp_in_sea_water
num_results: 0, result_count: 0
num_results: 0, result_count: 0
num_results: 0, result_count: 0, total_result_count: 12761
(12761, 4)
Do some summarizing of the responses#
The DataFrame of the matching datasets is quite large. I wonder what the distribution of those datasets across organizations looks like? Let’s use pandas.groupby() to generate some statistics about how many datasets are provided, matching our criteria, by which organization.
df_out.groupby(by="org").count()
| title | url | std_name | |
|---|---|---|---|
| org | |||
| AOOS | 28 | 28 | 28 |
| CeNCOOS | 416 | 416 | 416 |
| GCOOS | 9617 | 9617 | 9617 |
| Glider DAC | 2233 | 2233 | 2233 |
| MARACOOS | 221 | 221 | 221 |
| NANOOS | 4 | 4 | 4 |
| NERACOOS | 48 | 48 | 48 |
| PacIOOS | 16 | 16 | 16 |
| SCCOOS | 1 | 1 | 1 |
| SECOORA | 177 | 177 | 177 |
Drop the Glider DAC data#
Glider DAC data are already making it to NCEI, so we can drop those entries.
df_out_no_glider = df_out.loc[~df_out["org"].str.contains("Glider DAC")]
df_out_no_glider.groupby(by="org").count()
| title | url | std_name | |
|---|---|---|---|
| org | |||
| AOOS | 28 | 28 | 28 |
| CeNCOOS | 416 | 416 | 416 |
| GCOOS | 9617 | 9617 | 9617 |
| MARACOOS | 221 | 221 | 221 |
| NANOOS | 4 | 4 | 4 |
| NERACOOS | 48 | 48 | 48 |
| PacIOOS | 16 | 16 | 16 |
| SCCOOS | 1 | 1 | 1 |
| SECOORA | 177 | 177 | 177 |
Digging into some of the nuances#
There are still quite a lot of datasets from each organization. As our search above looked for each CF standard_name across all the datasets, there might be duplicate datasets which have multiple matching CF standard names. ie. one dataset might have both mass_concentration_of_oxygen_in_sea_water and fractional_saturation_of_oxygen_in_sea_water, but we only need to know that it’s one dataset.
As we only need to know about the unique datasets, let’s count how many unique dataset urls we have.
df_out_no_glider.groupby(by="url").count()
| title | org | std_name | |
|---|---|---|---|
| url | |||
| http://www.humboldt.edu | 4 | 4 | 4 |
| http://www.neracoos.org/erddap/tabledap/A01_aanderaa_o2_all | 1 | 1 | 1 |
| http://www.neracoos.org/erddap/tabledap/A01_optode_all | 2 | 2 | 2 |
| http://www.neracoos.org/erddap/tabledap/A01_sbe16_disox_all | 2 | 2 | 2 |
| http://www.neracoos.org/erddap/tabledap/C02_sbe16_disox_all | 2 | 2 | 2 |
| ... | ... | ... | ... |
| https://pae-paha.pacioos.hawaii.edu/erddap/tabledap/nss_012 | 2 | 2 | 2 |
| https://pae-paha.pacioos.hawaii.edu/erddap/tabledap/nss_013 | 2 | 2 | 2 |
| https://pae-paha.pacioos.hawaii.edu/erddap/tabledap/wqb_04 | 2 | 2 | 2 |
| https://pae-paha.pacioos.hawaii.edu/erddap/tabledap/wqb_05 | 2 | 2 | 2 |
| https://www.nodc.noaa.gov/ocads/oceans/time_series_moorings.html | 4 | 4 | 4 |
7654 rows × 3 columns
Drop duplicate records#
As you can see above, there are a lot of duplicate dataset urls which we can simplify down. We identify duplicates by looking at the URL, which should be unique for each dataset, and drop the duplicates.
df_out_nodups_no_glider = df_out_no_glider.drop_duplicates(subset=["url"], keep="last")
df_out_nodups_no_glider
| title | url | org | std_name | |
|---|---|---|---|---|
| 855 | Great Bay,NH. Oyster River WQ station | http://www.neracoos.org/erddap/tabledap/GRBORW... | NERACOOS | fractional_saturation_of_oxygen_in_sea_water |
| 857 | Great Bay,NH. Squamscott River WQ station | http://www.neracoos.org/erddap/tabledap/GRBSQW... | NERACOOS | fractional_saturation_of_oxygen_in_sea_water |
| 868 | Great Bay,NH. Great Bay WQ station | http://www.neracoos.org/erddap/tabledap/GRBGBW... | NERACOOS | fractional_saturation_of_oxygen_in_sea_water |
| 870 | Great Bay,NH. Lamprey River WQ station | http://www.neracoos.org/erddap/tabledap/GRBLRW... | NERACOOS | fractional_saturation_of_oxygen_in_sea_water |
| 891 | Monterey Bay Aquarium Seawater Intake | https://erddap.cencoos.org/erddap/tabledap/mon... | CeNCOOS | fractional_saturation_of_oxygen_in_sea_water |
| ... | ... | ... | ... | ... |
| 12756 | Walton-Smith CTD, WS1102, WS1102_028, 2011-02-... | https://gcoos5.geos.tamu.edu/erddap/tabledap/W... | GCOOS | volume_fraction_of_oxygen_in_sea_water |
| 12757 | Walton-Smith CTD, WS0718, WS0718_WS0718_056, 2... | https://gcoos5.geos.tamu.edu/erddap/tabledap/W... | GCOOS | volume_fraction_of_oxygen_in_sea_water |
| 12758 | Walton-Smith CTD, WS0802, WS0802_WS0802_Wet_40... | https://gcoos5.geos.tamu.edu/erddap/tabledap/W... | GCOOS | volume_fraction_of_oxygen_in_sea_water |
| 12759 | Walton-Smith CTD, WS20342, WS20342_WS20342_stn... | https://gcoos5.geos.tamu.edu/erddap/tabledap/W... | GCOOS | volume_fraction_of_oxygen_in_sea_water |
| 12760 | Walton-Smith CTD, WS22141, WS22141_WS22141_Stn... | https://gcoos5.geos.tamu.edu/erddap/tabledap/W... | GCOOS | volume_fraction_of_oxygen_in_sea_water |
7654 rows × 4 columns
How many endpoints are not ERDDAP?#
Now we have a unique list of datasets which match our CF standard name criteria. Since we have some background in using ERDDAP to query for data, let’s take a look at what other endpoints each of the datasets are using.
Hint: We know ERDDAP systems typically have erddap in their urls.
df_out_nodups_no_glider.loc[~df_out_nodups_no_glider["url"].str.contains("erddap")]
| title | url | org | std_name | |
|---|---|---|---|---|
| 9096 | CeNCOOS in situ water monitoring data at Trini... | http://www.humboldt.edu | CeNCOOS | mass_concentration_of_oxygen_in_sea_water |
| 9176 | MAPCO2 Buoy: Maihi Bay, Hawaii Island, Hawaii | https://www.nodc.noaa.gov/ocads/oceans/time_se... | PacIOOS | mass_concentration_of_oxygen_in_sea_water |
What’s the remaining distribution?#
This is the distribution of unique datasets found in the IOOS Data Catalog which have a CF Standard Name that contains the work oxygen and sea_water. We’ve dropped out the Glider DAC datasets as, theoretically, those are in NCEI already.
df_out_nodups_no_glider.groupby(by="org").count()
| title | url | std_name | |
|---|---|---|---|
| org | |||
| AOOS | 13 | 13 | 13 |
| CeNCOOS | 217 | 217 | 217 |
| GCOOS | 7152 | 7152 | 7152 |
| MARACOOS | 137 | 137 | 137 |
| NANOOS | 2 | 2 | 2 |
| NERACOOS | 32 | 32 | 32 |
| PacIOOS | 7 | 7 | 7 |
| SCCOOS | 1 | 1 | 1 |
| SECOORA | 93 | 93 | 93 |
Ingest data#
Let’s rip through all of the datasets, grab the data as a table (including units) and make a monster dictionary. This takes a bit.
import multiprocessing
from urllib.error import HTTPError
import joblib
import stamina
from tqdm import tqdm
@stamina.retry(on=HTTPError, attempts=3)
def request_df(url):
"""Thin layer to handle retries."""
return pd.read_csv(url, low_memory=False)
def error_handling_layer(row):
"""Even with stamina we may hit servers that will fail."""
title = row["title"]
# Requesting only the position.
url = f"{row['url']}.csvp?latitude,longitude&distinct()"
try:
df = request_df(url)
except Exception as err:
msg = f"Failed to fetch {url}. {err}."
print(msg)
df = None
return title, df
n_iter = len(df_out_nodups_no_glider)
num_cores = multiprocessing.cpu_count()
downloads = [
r
for r in tqdm(
joblib.Parallel(return_as="generator", n_jobs=num_cores, max_nbytes=5000)(
joblib.delayed(error_handling_layer)(row)
for _, row in df_out_nodups_no_glider.iterrows()
),
total=n_iter,
)
]
dict_out_final = dict(downloads)
0%| | 1/7654 [00:01<2:15:29, 1.06s/it]stamina.retry_scheduled
stamina.retry_scheduled
22%|█████████████████████████████▋ | 1706/7654 [01:33<16:15, 6.10it/s]stamina.retry_scheduled
stamina.retry_scheduled
23%|███████████████████████████████ | 1786/7654 [02:12<39:54, 2.45it/s]stamina.retry_scheduled
stamina.retry_scheduled
25%|█████████████████████████████████▏ | 1911/7654 [02:44<20:35, 4.65it/s]stamina.retry_scheduled
stamina.retry_scheduled
70%|████████████████████████████████████████████████████████████████████████████████████████████▊ | 5344/7654 [05:50<02:54, 13.21it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████ | 5359/7654 [06:10<30:29, 1.25it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▎ | 5371/7654 [06:29<30:52, 1.23it/s]stamina.retry_scheduled
stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▎ | 5372/7654 [06:31<35:42, 1.07it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
70%|███████████████████████████████████████████████████████████████████████████████████████████▉ | 5373/7654 [06:42<2:03:47, 3.26s/it]stamina.retry_scheduled
stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▍ | 5379/7654 [06:47<46:42, 1.23s/it]stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▌ | 5383/7654 [06:49<30:15, 1.25it/s]stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▌ | 5384/7654 [06:50<32:27, 1.17it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
70%|████████████████████████████████████████████████████████████████████████████████████████████▏ | 5385/7654 [07:01<1:53:10, 2.99s/it]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▋ | 5392/7654 [07:07<35:12, 1.07it/s]stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▋ | 5395/7654 [07:09<29:00, 1.30it/s]stamina.retry_scheduled
70%|█████████████████████████████████████████████████████████████████████████████████████████████▊ | 5396/7654 [07:10<34:15, 1.10it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
71%|████████████████████████████████████████████████████████████████████████████████████████████▎ | 5397/7654 [07:20<2:18:04, 3.67s/it]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
71%|█████████████████████████████████████████████████████████████████████████████████████████████▉ | 5404/7654 [07:26<36:53, 1.02it/s]stamina.retry_scheduled
71%|█████████████████████████████████████████████████████████████████████████████████████████████▉ | 5407/7654 [07:29<30:13, 1.24it/s]stamina.retry_scheduled
71%|█████████████████████████████████████████████████████████████████████████████████████████████▉ | 5408/7654 [07:30<32:02, 1.17it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
71%|████████████████████████████████████████████████████████████████████████████████████████████▌ | 5409/7654 [07:40<1:52:56, 3.02s/it]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
71%|████████████████████████████████████████████████████████████████████████████████████████████▌ | 5410/7654 [07:42<1:44:49, 2.80s/it]stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████ | 5414/7654 [07:45<49:45, 1.33s/it]stamina.retry_scheduled
stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████▏ | 5417/7654 [07:47<39:10, 1.05s/it]stamina.retry_scheduled
stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████▏ | 5419/7654 [07:48<32:11, 1.16it/s]stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████▏ | 5420/7654 [07:49<34:28, 1.08it/s]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████▏ | 5421/7654 [07:53<58:48, 1.58s/it]stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████▏ | 5423/7654 [07:54<40:47, 1.10s/it]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
71%|██████████████████████████████████████████████████████████████████████████████████████████████▎ | 5424/7654 [07:55<38:10, 1.03s/it]stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
stamina.retry_scheduled
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 7654/7654 [12:19<00:00, 10.35it/s]
Let’s take a quick look at one of the DataFrames.
Transpose it when we print, so we can see all the columns.
dict_out_final['"Deepwater CTD - pe972218.ctd.nc - 29.25N, -87.89W - 1997-03-21"'].head(
5
).T
| 0 | |
|---|---|
| latitude (degrees_north) | 29.247200 |
| longitude (degrees_east) | -87.888901 |
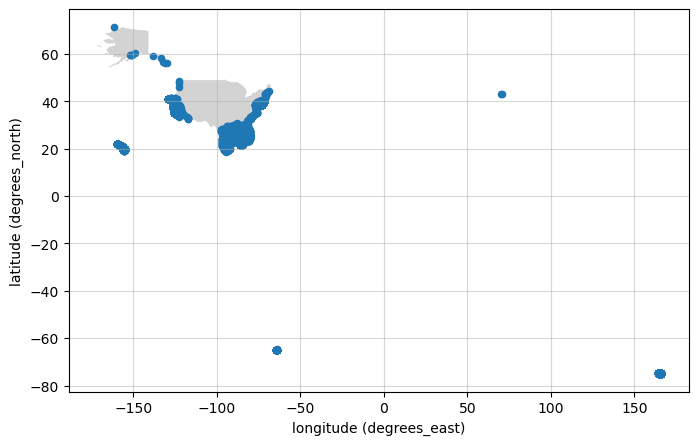
Let’s make a nice map of the distribution of observations#
Below we create a mapping function to plot the unique dataset points on a map. Then, we use that function with our full response. We have to do a little reorganizing of the data to build one DataFrame for all the coordinates.
import cartopy.io.shapereader as shpreader
import geopandas as gpd
import matplotlib.pyplot as plt
def make_map(df):
fig, ax = plt.subplots(figsize=(8, 6))
shpfilename = shpreader.natural_earth(
resolution="110m",
category="cultural",
name="admin_0_countries",
)
countries = gpd.read_file(shpfilename)
countries[countries["NAME"] == "United States of America"].plot(
color="lightgrey", ax=ax
)
df.plot(
x="longitude (degrees_east)",
y="latitude (degrees_north)",
kind="scatter",
ax=ax,
)
ax.grid(visible=True, alpha=0.5)
return ax
df_coords_clean = pd.concat(dict_out_final).dropna().drop_duplicates(ignore_index=True)
# Clean value outside of valid lon, lat.
df_coords_clean = df_coords_clean.loc[df_coords_clean["latitude (degrees_north)"] <= 90]
df_coords_clean = df_coords_clean.loc[
df_coords_clean["latitude (degrees_north)"] >= -90
]
df_coords_clean = df_coords_clean.loc[
df_coords_clean["longitude (degrees_east)"] <= 180
]
df_coords_clean = df_coords_clean.loc[
df_coords_clean["longitude (degrees_east)"] >= -180
]
df_coords_clean
make_map(df_coords_clean)
<Axes: xlabel='longitude (degrees_east)', ylabel='latitude (degrees_north)'>
Lets explore those points on an interactive map#
Just for fun, we can us folium’s MarkerCluster to plot many points on an interactive map to browse around. We have +7k points, marker cluster is quite robust but that many point can crash the browser, so let’s limit to the first 1k.
locations = list(
zip(
df_coords_clean["latitude (degrees_north)"][0:1000],
df_coords_clean["longitude (degrees_east)"][0:1000],
)
)
import folium
from folium.plugins import MarkerCluster
m = folium.Map(zoom_start=5)
marker_cluster = MarkerCluster().add_to(m)
icon_create_function = """\
function(cluster) {
return L.divIcon({
html: '<b>' + cluster.getChildCount() + '</b>',
className: 'marker-cluster marker-cluster-large',
iconSize: new L.Point(20, 20)
});
}"""
marker_cluster = MarkerCluster(
locations=locations,
overlay=True,
control=True,
icon_create_function=icon_create_function,
)
marker_cluster.add_to(m)
folium.LayerControl().add_to(m)
m.fit_bounds(m.get_bounds())
m
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1988.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1989.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1994.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1977.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2005.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1983.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2006.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1997.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1988.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_2002.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1986.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_2005.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1992.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.aoos.org/erddap/tabledap/homer-dolphin-surface-water-q.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1998.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_2000.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1996.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_2002.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1985.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1996.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1993.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/monterey-bay-aquarium-seawate.csvp?latitude,longitude&distinct(). HTTP Error 500: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2000.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1984.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_2005.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2002.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1975.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1983.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_elkapwq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1989.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1990.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1987.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_2003.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_2001.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1986.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_elkvmwq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1980.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1987.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1974.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1995.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_2003.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1992.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/monterey-wharf-real-time-samplin.csvp?latitude,longitude&distinct(). HTTP Error 500: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1999.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1999.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1982.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1994.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2004.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1993.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://www.nodc.noaa.gov/ocads/oceans/time_series_moorings.html.csvp?latitude,longitude&distinct(). HTTP Error 404: Not Found.
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_sfbccwq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/mlml_monterey.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1997.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1998.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1981.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_2004.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2003.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_2001.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1976.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_elksmwq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/morro-bay-bs1.csvp?latitude,longitude&distinct(). HTTP Error 500: .
Failed to fetch https://erddap.secoora.org/erddap/tabledap/pivers-island-coastal-observa.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1990.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_2000.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1991.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1981.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1978.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_2004.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_elknmwq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1990.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1997.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1991.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1984.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1982.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1995.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_tjroswq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch http://www.humboldt.edu.csvp?latitude,longitude&distinct(). <urlopen error [Errno -2] Name or service not known>.
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_1998.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1973.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1991.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1979.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1994.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1992.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1996.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1999.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2007.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1985.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Alabama_CPUE_Trawl_study_DATA_2001.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Florida_CPUE_Trawl_study_DATA_1995.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos4.geos.tamu.edu/erddap/tabledap/CAGES_Mississippi_CPUE_Trawl_study_DATA_1993.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://erddap.cencoos.org/erddap/tabledap/nerrs_tjrsbwq.csvp?latitude,longitude&distinct(). HTTP Error 404: .
Failed to fetch https://gcoos5.geos.tamu.edu/erddap/tabledap/WS1202_WS1202_deck-tst.csvp?latitude,longitude&distinct(). <urlopen error [Errno 110] Connection timed out>.
We hope this example demonstrates the flexibility of direct requests to the IOOS Data Catalog CKAN server and all the possibilities it provides. In this notebook we:
Search the IOOS Data Catalog CKAN API with keywords.
Found datasets matching our specified criteria.
Collected all the data from each of the datasets matching our criteria.
Created a simple map of the distribution of datasets which match our criteria.
To take this one step further, since we collected all the data from each of the datasets (in the dictionary dict_out_final) a user could integrate all of the oxygen observations together and start to build a comprehensive dataset.
Additionally, a user could modify the CKAN query to search for terms outside of the CF standard names to potentially gather more datasets.


