Interpolate MURSST to great white shark telemetry track with R and ERDDAP#
Created: 2024-05-16
Updated: 2024-07-02
This notebook walks through downloading a netCDF file from NCEI. The file follows a specific specification for ATN satellite trajectory observations as documented here. More information about the ATN netCDF specification can be found at https://ioos.github.io/ioos-atn-data/.
Since most of the examples in the IOOS Code Lab are in the python programming language, we wanted to show an example of working with a netCDF file in the R programming language to be inclusive of those users.
For more information on the tidync package, see this R blog: https://ropensci.org/blog/2019/11/05/tidync/
Data used in this notebook are available from NCEI at the following link https://www.ncei.noaa.gov/archive/accession/0282699.
library(tidync)
library(obistools)
library(ncdf4)
library(tidyverse)
library(lubridate)
library(maps)
library(mapdata)
The legacy packages maptools, rgdal, and rgeos, underpinning the sp package,
which was just loaded, will retire in October 2023.
Please refer to R-spatial evolution reports for details, especially
https://r-spatial.org/r/2023/05/15/evolution4.html.
It may be desirable to make the sf package available;
package maintainers should consider adding sf to Suggests:.
The sp package is now running under evolution status 2
(status 2 uses the sf package in place of rgdal)
-- Attaching packages ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- tidyverse 1.3.2 --
v ggplot2 3.4.2 v purrr 1.0.1
v tibble 3.2.1 v dplyr 1.1.1
v tidyr 1.3.0 v stringr 1.5.0
v readr 2.1.4 v forcats 1.0.0
-- Conflicts -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- tidyverse_conflicts() --
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()
Attaching package: 'lubridate'
The following objects are masked from 'package:base':
date, intersect, setdiff, union
Attaching package: 'maps'
The following object is masked from 'package:purrr':
map
Download the netCDF file from the NCEI Archive#
See https://www.ncei.noaa.gov/archive/accession/0282699
url = 'https://www.ncei.noaa.gov/data/oceans/archive/arc0217/0282699/1.1/data/0-data/atn_45866_great-white-shark_trajectory_20090923-20091123.nc'
fname = basename(url)
download.file(url, fname, mode = "wb")
Collect all the metadata from the netCDF file.#
This gathers not only the global attributes, but the variable level attributes as well. As you can see in the variable column the term NC_GLOBAL refers to global attributes.
metadata <- ncmeta::nc_atts(fname)
Example extracting attributes from netCDF#
To give you an idea of what observations are in this file, we can print out the human readable summary and long_name attributes from all the variables.
metadata %>% dplyr::filter(name == "summary")
| id | name | variable | value |
|---|---|---|---|
| <int> | <chr> | <chr> | <named list> |
| 46 | summary | NC_GLOBAL | Wildlife Computers SPOT5 tag (ptt id 45866) deployed on a great white shark (Carcharodon carcharias) by Chris G. Lowe in the North Pacific Ocean from 2009-09-23 to 2009-11-23 |
metadata %>% dplyr::filter(name == "long_name")
| id | name | variable | value |
|---|---|---|---|
| <int> | <chr> | <chr> | <named list> |
| 0 | long_name | deploy_id | id for this deployment. This is typically the tag ptt |
| 5 | long_name | time | Time of the measurement, in seconds since 1990-01-01 |
| 2 | long_name | z | depth of measurement |
| 2 | long_name | lat | Latitude portion of location in decimal degrees North |
| 2 | long_name | lon | Longitude portion of location in decimal degrees East |
| 2 | long_name | ptt | Platform Transmitter Terminal (PTT) id used for Argos transmissions |
| 2 | long_name | instrument | Instrument family |
| 2 | long_name | type | Type of location information - Argos, GPS satellite or user provided location |
| 3 | long_name | location_class | Location Quality Code from ARGOS satellite system |
| 2 | long_name | error_radius | Error radius |
| 2 | long_name | semi_major_axis | Error - ellipse semi-major axis |
| 2 | long_name | semi_minor_axis | Error - ellipse semi-minor axis |
| 2 | long_name | ellipse_orientation | Error - ellipse orientation in degrees clockwise from true north |
| 2 | long_name | offset | Error - offset in meters to center of error ellipse or circle |
| 2 | long_name | offset_orientation | Error - offset orientation angle to ellipse center |
| 2 | long_name | gpe_msd | |
| 2 | long_name | gpe_u | |
| 3 | long_name | count | Count |
| 3 | long_name | qartod_time_flag | Time QC test - gross range test |
| 3 | long_name | qartod_speed_flag | Speed QC test - gross range test |
| 3 | long_name | qartod_location_flag | Location QC test - Location test |
| 3 | long_name | qartod_rollup_flag | Aggregate QC value |
| 3 | long_name | crs | Coordinate Reference System - http://www.opengis.net/def/crs/EPSG/0/4326 |
| 1 | long_name | trajectory | trajectory identifier |
| 2 | long_name | animal_age | age of the animal as measured or estimated at deployment |
| 1 | long_name | animal_life_stage | Lifestage of the animal at time of deployment |
| 1 | long_name | animal_sex | sex of the animal at time of tag deployment |
| 2 | long_name | animal_weight | mass of the animal as measured or estimated at deployment |
| 4 | long_name | animal_length | length of the animal as measured or estimated at deployment |
| 4 | long_name | animal_length_2 | length of the animal as measured or estimated at deployment |
| 3 | long_name | animal | tagged animal id |
| 3 | long_name | instrument_tag | telemetry tag applied to animal |
| 3 | long_name | instrument_location | Wildlife Computers SPOT5 |
| 1 | long_name | taxon_name | most precise taxonomic classification for the tagged animal |
| 1 | long_name | taxon_lsid | Namespaced Taxon Identifier for the tagged animal |
| 0 | long_name | comment | Comment |
Store the data as a tibble in a variable#
Collect the data dimensioned by time from the netCDF file as a tibble. Then, print the first few rows.
atn <- tidync(fname)
atn_tbl <- atn %>% hyper_tibble(force=TRUE)
head(atn_tbl, n=4)
| time | z | lat | lon | ptt | instrument | type | location_class | error_radius | semi_major_axis | ... | offset_orientation | gpe_msd | gpe_u | count | qartod_time_flag | qartod_speed_flag | qartod_location_flag | qartod_rollup_flag | comment | obs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <dbl> | <int> | <dbl> | <dbl> | <int> | <chr> | <chr> | <chr> | <int> | <int> | ... | <int> | <dbl> | <dbl> | <int> | <int> | <int> | <int> | <int> | <chr> | <int> |
| 622512000 | 0 | 34.030 | -118.560 | 45866 | SPOT | User | nan | NA | NA | ... | NA | NaN | NaN | NA | 1 | 2 | 1 | 1 | 1 | |
| 622708920 | 0 | 23.590 | -166.180 | 45866 | SPOT | Argos | A | NA | NA | ... | NA | NaN | NaN | NA | 1 | 4 | 1 | 4 | 2 | |
| 622724940 | 0 | 34.024 | -118.556 | 45866 | SPOT | Argos | 1 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 4 | 1 | 4 | 3 | |
| 622725060 | 0 | 34.035 | -118.549 | 45866 | SPOT | Argos | 0 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 4 | 1 | 4 | 4 |
Dealing with time#
Notice the data in the time column aren’t formatted as times. We need to read the metadata associated with the time variable to understand what the units are. Below, we print a tibble of all the attributes from the time variable.
Notice the units attribute and it’s value of seconds since 1990-01-01 00:00:00Z. We need to use that information to convert the time variable to something useful that ggplot can handle.
time_attrs <- metadata %>% dplyr::filter(variable == "time")
time_attrs
| id | name | variable | value |
|---|---|---|---|
| <int> | <chr> | <chr> | <named list> |
| 0 | units | time | seconds since 1990-01-01 00:00:00Z |
| 1 | standard_name | time | time |
| 2 | axis | time | T |
| 3 | _CoordinateAxisType | time | Time |
| 4 | calendar | time | standard |
| 5 | long_name | time | Time of the measurement, in seconds since 1990-01-01 |
| 6 | actual_min | time | 2009-09-23T00:00:00Z |
| 7 | actual_max | time | 2009-11-23T05:12:00Z |
| 8 | ancillary_variables | time | qartod_time_flag qartod_rollup_flag qartod_speed_flag |
| 9 | instrument | time | instrument_location |
| 10 | platform | time | animal |
| 11 | coverage_content_type | time | coordinate |
| 12 | _FillValue | time | NaN |
So, we grab the value from the units attribute, split the string to collect the date information, and apply that to a time conversion function as.POSIXct.
tunit <- time_attrs %>% dplyr::filter(name == "units")
lunit <- str_split(tunit$value,' ')[[1]]
atn_tbl$time <- as.POSIXct(atn_tbl$time, origin=lunit[3], tz="GMT")
head(atn_tbl, n=2)
| time | z | lat | lon | ptt | instrument | type | location_class | error_radius | semi_major_axis | ... | offset_orientation | gpe_msd | gpe_u | count | qartod_time_flag | qartod_speed_flag | qartod_location_flag | qartod_rollup_flag | comment | obs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <dttm> | <int> | <dbl> | <dbl> | <int> | <chr> | <chr> | <chr> | <int> | <int> | ... | <int> | <dbl> | <dbl> | <int> | <int> | <int> | <int> | <int> | <chr> | <int> |
| 2009-09-23 00:00:00 | 0 | 34.03 | -118.56 | 45866 | SPOT | User | nan | NA | NA | ... | NA | NaN | NaN | NA | 1 | 2 | 1 | 1 | 1 | |
| 2009-09-25 06:42:00 | 0 | 23.59 | -166.18 | 45866 | SPOT | Argos | A | NA | NA | ... | NA | NaN | NaN | NA | 1 | 4 | 1 | 4 | 2 |
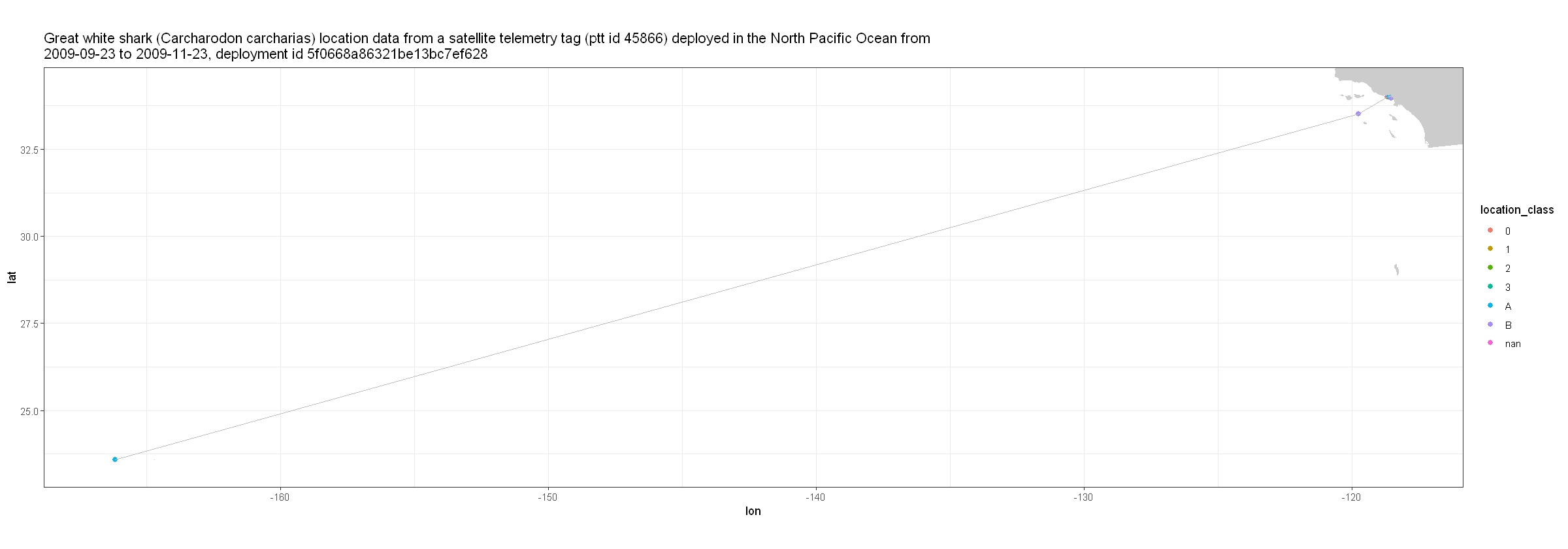
Plotting the raw data#
Now lets make some plots just to take a look at what data we’re working with. Here we make a plot of time vs longitude.
options(repr.plot.width = 20, repr.plot.height = 7)
# collect title from global attributes
title <- metadata %>% dplyr::filter(variable == "NC_GLOBAL") %>% dplyr::filter(name == "title")
# Map limits.
ylim <- c( min(atn_tbl$lat)-0.25, max(atn_tbl$lat)+0.25 )
xlim <- c( min(atn_tbl$lon)-0.25, max(atn_tbl$lon)+0.25 )
# Get outline data for map.
w <- map_data( 'worldHires', ylim = ylim, xlim = xlim )
z <- ggplot(atn_tbl, aes( x = lon, y = lat )) +
geom_point( aes(color=location_class), size=2) +
geom_line( color = 'grey' ) +
scale_shape_manual( values = c(19, 1) )
z + geom_polygon(data = w, aes(x = long, y = lat, group = group), fill = 'grey80') +
theme_bw() +
coord_fixed(1.3, xlim = xlim, ylim = ylim) +
labs(title = paste( strwrap(title$value, width = 150), collapse = "\n"))
What do these location_class codes mean?
code_defs <- metadata %>% dplyr::filter(variable == "location_class") %>% dplyr::filter(name=="code_meanings" | name=="code_values")
str_split_fixed(code_defs$value,",",8)
| G | 3 | 2 | 1 | 0 | A | B | Z |
| estimated error less than 100m and 1+ messages received per satellite pass | estimated error less than 250m and 4+ messages received per satellite pass | estimated error between 250m and 500m and 4+ messages per satellite pass | estimated error between 500m and 1500m and 4+ messages per satellite pass | estimated error greater than 1500m and 4+ messages received per satellite pass | no least squares estimated error or unbounded kalman filter estimated error and 3 messages received per satellite pass | no least squares estimated error or unbounded kalman filter estimated error and 1 or 2 messages received per satellite pass | invalid location (available for Service Plus or Auxilliary Location Processing) |
Review location uncertainty information and map to how many meters the uncertainty is.
From the table above, only nan, G, 3, 2, 1, and 0 are valid points with known undertainties.
atn_tbl_qc <- atn_tbl %>%
filter(location_class %in% c('nan','G','3','2','1','0')) %>%
# mutate( # This returns NA for any other values than those defined below
# coordinateUncertaintyInMeters = case_when(location_class == 'nan' ~ 0,
# location_class == 'G' ~ 200,
# location_class == '3' ~ 250,
# location_class == '2' ~ 500,
# location_class == '1' ~ 1500,
# location_class == '0' ~ 10000) # https://github.com/ioos/bio_data_guide/issues/145#issuecomment-1805739244
# ) %>%
arrange(time)
head(atn_tbl_qc)
| time | z | lat | lon | ptt | instrument | type | location_class | error_radius | semi_major_axis | ... | offset_orientation | gpe_msd | gpe_u | count | qartod_time_flag | qartod_speed_flag | qartod_location_flag | qartod_rollup_flag | comment | obs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <dttm> | <int> | <dbl> | <dbl> | <int> | <chr> | <chr> | <chr> | <int> | <int> | ... | <int> | <dbl> | <dbl> | <int> | <int> | <int> | <int> | <int> | <chr> | <int> |
| 2009-09-23 00:00:00 | 0 | 34.030 | -118.560 | 45866 | SPOT | User | nan | NA | NA | ... | NA | NaN | NaN | NA | 1 | 2 | 1 | 1 | 1 | |
| 2009-09-25 11:09:00 | 0 | 34.024 | -118.556 | 45866 | SPOT | Argos | 1 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 4 | 1 | 4 | 3 | |
| 2009-09-25 11:11:00 | 0 | 34.035 | -118.549 | 45866 | SPOT | Argos | 0 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 4 | 1 | 4 | 4 | |
| 2009-09-27 17:58:00 | 0 | 34.033 | -118.547 | 45866 | SPOT | Argos | 1 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 1 | 1 | 1 | 5 | |
| 2009-10-08 20:24:00 | 0 | 34.038 | -118.581 | 45866 | SPOT | Argos | 2 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 1 | 1 | 1 | 7 | |
| 2009-10-15 11:05:00 | 0 | 33.995 | -118.678 | 45866 | SPOT | Argos | 0 | NA | NA | ... | NA | NaN | NaN | NA | 1 | 1 | 1 | 1 | 8 |
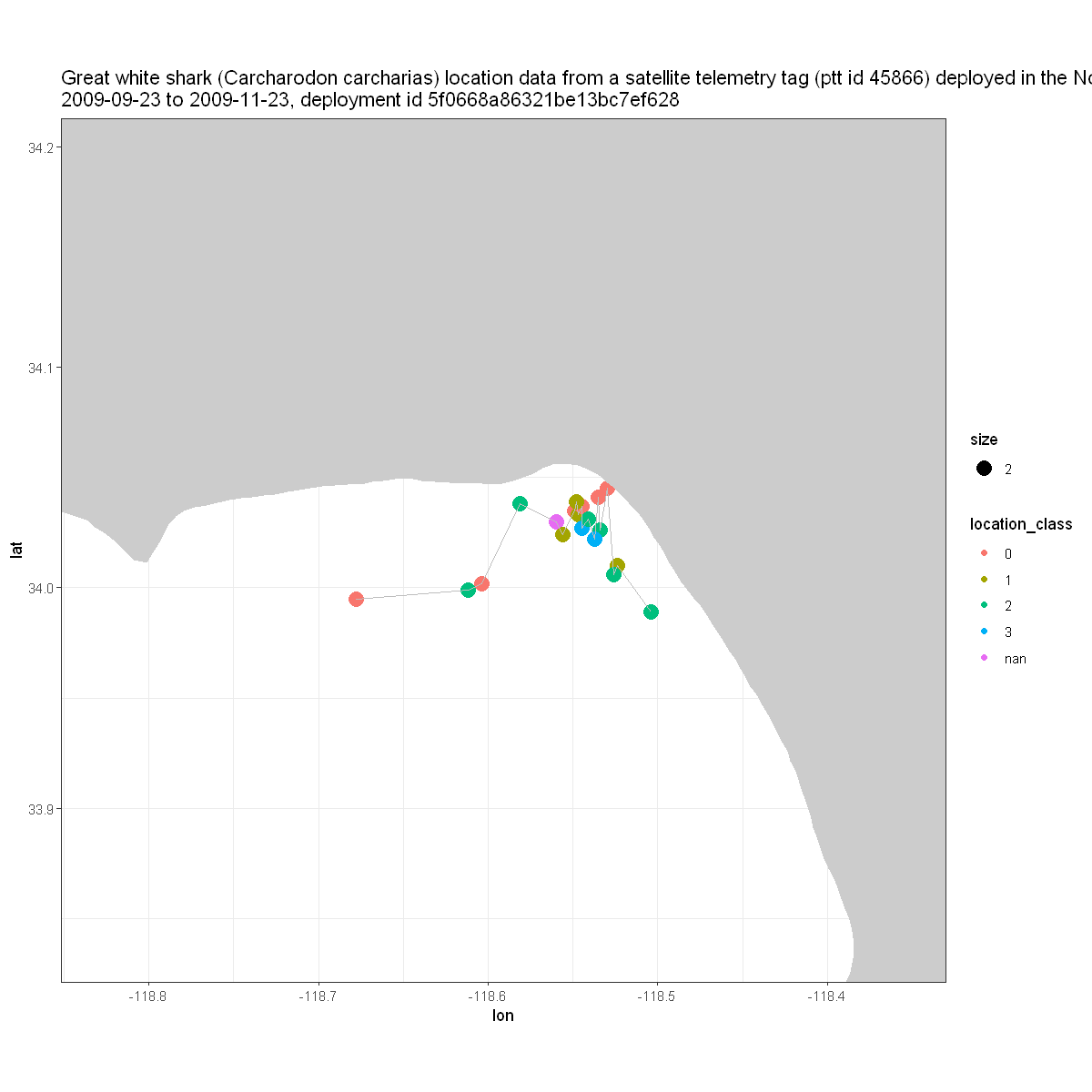
Making a map#
Next, let’s make a map of the locations of this animal with the colors identifying the location classes described in the table above.
options(repr.plot.width = 10, repr.plot.height = 10)
# collect title from global attributes
title <- metadata %>% dplyr::filter(variable == "NC_GLOBAL") %>% dplyr::filter(name == "title")
# Map limits.
ylim <- c( min(atn_tbl_qc$lat)-0.15, max(atn_tbl_qc$lat)+0.15 )
xlim <- c( min(atn_tbl_qc$lon)-0.15, max(atn_tbl_qc$lon)+0.15 )
# Get outline data for map.
w <- map_data( 'worldHires', ylim = ylim, xlim = xlim )
z <- ggplot(atn_tbl_qc, aes( x = lon, y = lat )) +
geom_point( aes(color = location_class, size = 2 )) +
geom_line( color='grey' ) +
scale_shape_manual( values = c(19, 1) )
z + geom_polygon(data = w, aes(x = long, y = lat, group = group), fill = 'grey80') +
theme_bw() +
coord_fixed(1.3, xlim = xlim, ylim = ylim) +
labs(title = paste( strwrap(title$value, width = 150), collapse = "\n"))
Interpolating MURSST to white shark tracks using ERDDAP#
The MURSST product is a merged, multi-sensor L4 Foundation Sea Surface Temperature (SST) analysis product from Jet Propulsion Laboratory (JPL). This daily, global, Multi-scale, Ultra-high Resolution (MUR) Sea Surface Temperature (SST) 1-km data set, Version 4.1, is produced at JPL under the NASA MEaSUREs program. For details, see https://podaac.jpl.nasa.gov/dataset/MUR-JPL-L4-GLOB-v4.1. This dataset is part of the Group for High-Resolution Sea Surface Temperature (GHRSST) project. The data for the most recent 7 days is usually revised everyday. The data for other days is sometimes revised.
We need to prepare the data to send to the server. As mentioned above the request will take a table with a header "time,latitude,longitude", then its values. Note that we rename the columns, then pick the subset to store as one long string to create the request.
time_lat_lon <- atn_tbl_qc %>%
rename(
latitude = lat,
longitude = lon)
time_lat_lon <- format_csv(time_lat_lon[c('time','latitude','longitude')])
The cell below builds the request and download get it back as a data.frame.
response = "csv"
dataset_id = "jplMURSST41"
variable = "analysed_sst"
algorithm = "Nearest"
nearby = 216
url = paste0(
"http://coastwatch.pfeg.noaa.gov/erddap/",
"convert/interpolate.",response,
"?TimeLatLonTable=",URLencode(time_lat_lon, reserved=TRUE),
"&requestCSV=",dataset_id,
"/",variable,
"/",algorithm,
"/",nearby
)
df <- read.csv(url)
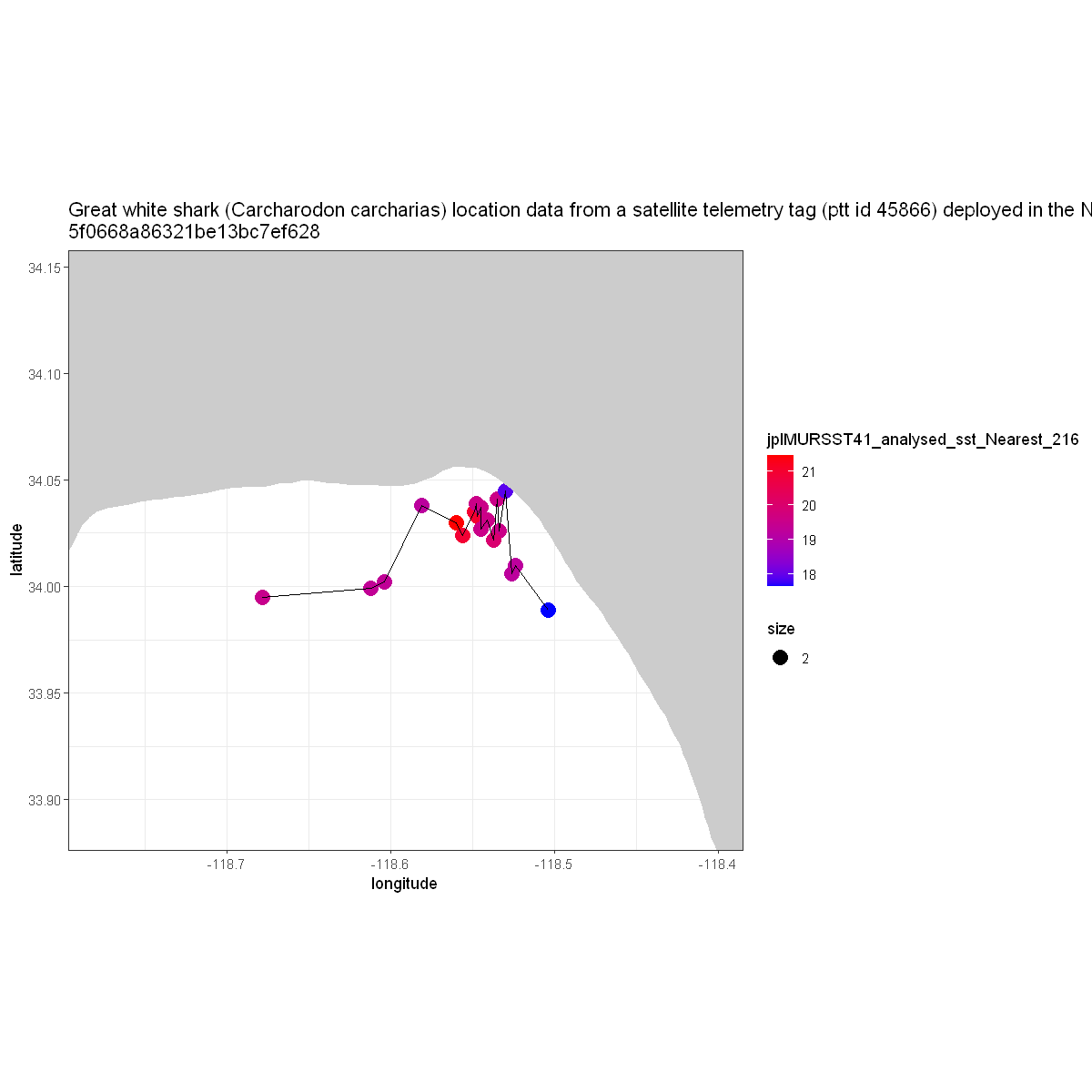
Now let’s plot what the interpolated sea surface temperatures were for that animal at that location and time.
options(repr.plot.width = 10, repr.plot.height = 10)
# collect title from global attributes
title <- metadata %>% dplyr::filter(variable == "NC_GLOBAL") %>% dplyr::filter(name == "title")
# Map limits.
ylim <- c( min(df$latitude)-0.1, max(df$latitude)+0.1 )
xlim <- c( min(df$longitude)-0.1, max(df$longitude)+0.1 )
# Get outline data for map.
w <- map_data( 'worldHires', ylim = ylim, xlim = xlim )
z <- ggplot(df, aes( x = longitude, y = latitude )) +
geom_point( aes(color = jplMURSST41_analysed_sst_Nearest_216, size=2)) +
geom_line() +
scale_shape_manual( values = c(22, 17) ) +
scale_color_gradient(low="blue", high="red")
z + geom_polygon(data = w, aes(x = long, y = lat, group = group), fill = 'grey80') +
theme_bw() +
coord_fixed(1.3, xlim = xlim, ylim = ylim) +
labs(title = paste( strwrap(title$value, width = 200), collapse = "\n"))
sessionInfo()
R version 4.1.3 (2022-03-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mapdata_2.3.1 maps_3.4.1 lubridate_1.9.2 forcats_1.0.0
[5] stringr_1.5.0 dplyr_1.1.1 purrr_1.0.1 readr_2.1.4
[9] tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.2 tidyverse_1.3.2
[13] ncdf4_1.21 obistools_0.0.10 tidync_0.3.0
loaded via a namespace (and not attached):
[1] httr_1.4.6 bit64_4.0.5 vroom_1.6.1
[4] jsonlite_1.8.4 modelr_0.1.11 sp_2.0-0
[7] googlesheets4_1.1.0 cellranger_1.1.0 pillar_1.9.0
[10] backports_1.4.1 lattice_0.20-45 glue_1.6.2
[13] uuid_1.1-0 digest_0.6.31 rvest_1.0.3
[16] colorspace_2.1-0 htmltools_0.5.5 pkgconfig_2.0.3
[19] broom_1.0.4 haven_2.5.2 scales_1.2.1
[22] tzdb_0.3.0 timechange_0.2.0 googledrive_2.1.0
[25] farver_2.1.1 generics_0.1.3 withr_2.5.0
[28] repr_1.1.6 cli_3.6.0 readxl_1.4.2
[31] magrittr_2.0.3 crayon_1.5.2 evaluate_0.20
[34] data.tree_1.0.0 fs_1.6.1 fansi_1.0.4
[37] xml2_1.3.3 tools_4.1.3 hms_1.1.3
[40] geosphere_1.5-18 gargle_1.4.0 lifecycle_1.0.3
[43] reprex_2.0.2 munsell_0.5.0 compiler_4.1.3
[46] RNetCDF_2.6-2 rlang_1.1.0 grid_4.1.3
[49] pbdZMQ_0.3-9 IRkernel_1.3.2 rappdirs_0.3.3
[52] htmlwidgets_1.6.2 crosstalk_1.2.0 labeling_0.4.2
[55] base64enc_0.1-3 rmarkdown_2.20 gtable_0.3.3
[58] DBI_1.1.3 R6_2.5.1 ncmeta_0.3.5
[61] knitr_1.42 bit_4.0.5 fastmap_1.1.1
[64] rgeos_0.6-2 utf8_1.2.3 stringi_1.7.12
[67] parallel_4.1.3 IRdisplay_1.1 Rcpp_1.0.10
[70] vctrs_0.6.1 leaflet_2.1.2 dbplyr_2.3.2
[73] tidyselect_1.2.0 xfun_0.37