Quick demonstration of R-notebooks using the r-oce library#
Created: 2017-01-23
The IOOS notebook
environment
installs the R language and the Jupyter kernel needed to run R notebooks.
Conda can also install extra R packages,
and those packages that are unavailable in conda can be installed directly from CRAN with install.packages(pkg_name).
You can start jupyter from any other environment and change the kernel later using the drop-down menu.
(Check the R logo at the top right to ensure you are in the R jupyter kernel.)
In this simple example we will use two libraries aimed at the oceanography community written in R: r-gsw and r-oce.
(The original post for the examples below can be found author’s blog: https://dankelley.github.io/blog/)
library(gsw)
library(oce)
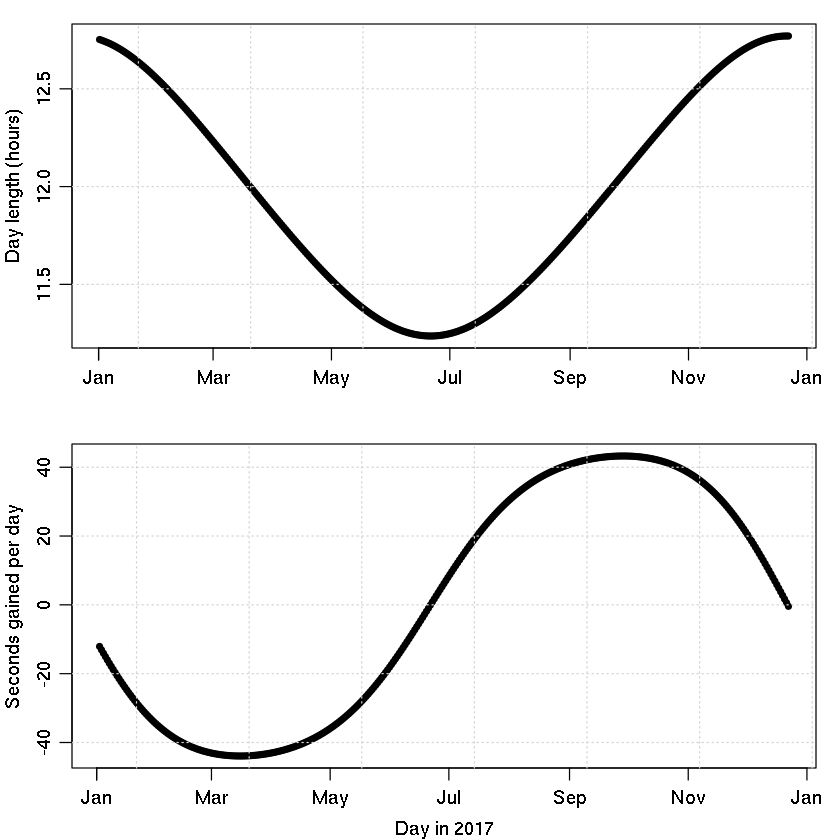
Example 1: calculating the day length.
daylength <- function(t, lon=-38.5, lat=-13)
{
t <- as.numeric(t)
alt <- function(t)
sunAngle(t, longitude=lon, latitude=lat)$altitude
rise <- uniroot(alt, lower=t-86400/2, upper=t)$root
set <- uniroot(alt, lower=t, upper=t+86400/2)$root
set - rise
}
t0 <- as.POSIXct("2017-01-01 12:00:00", tz="UTC")
t <- seq.POSIXt(t0, by="1 day", length.out=1*356)
dayLength <- unlist(lapply(t, daylength))
par(mfrow=c(2,1), mar=c(3, 3, 1, 1), mgp=c(2, 0.7, 0))
plot(t, dayLength/3600, type='o', pch=20,
xlab="", ylab="Day length (hours)")
grid()
solstice <- as.POSIXct("2013-12-21", tz="UTC")
plot(t[-1], diff(dayLength), type='o', pch=20,
xlab="Day in 2017", ylab="Seconds gained per day")
grid()
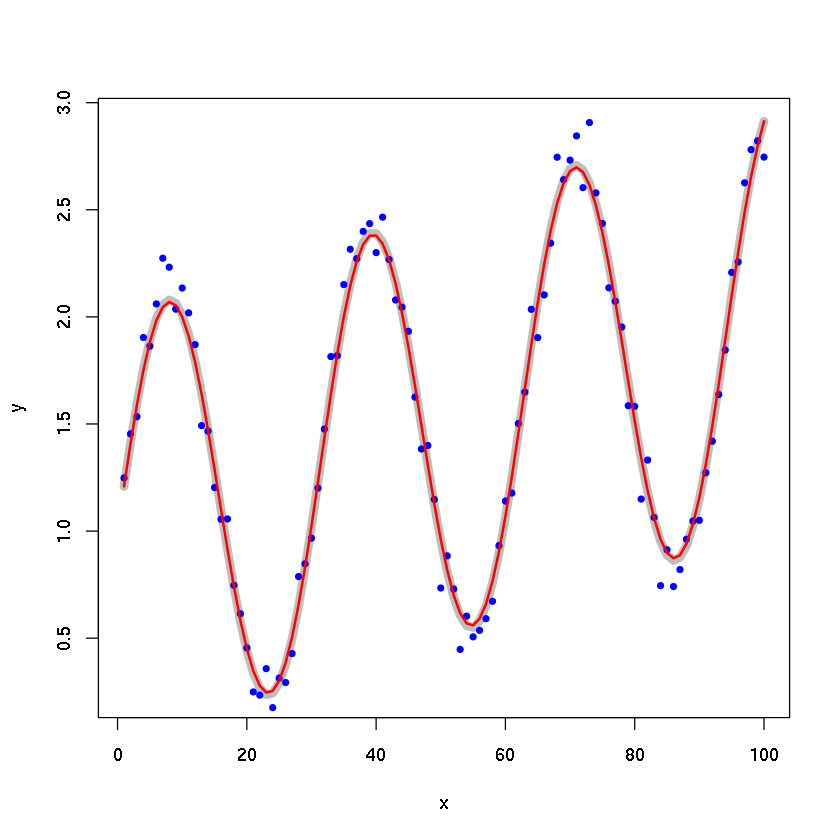
Example 2: least-square fit.
x <- 1:100
y <- 1 + x/100 + sin(x/5)
yn <- y + rnorm(100, sd=0.1)
L <- 4
calc <- runlm(x, y, L=L, deriv=0)
plot(x, y, type='l', lwd=7, col='gray')
points(x, yn, pch=20, col='blue')
lines(x, calc, lwd=2, col='red')
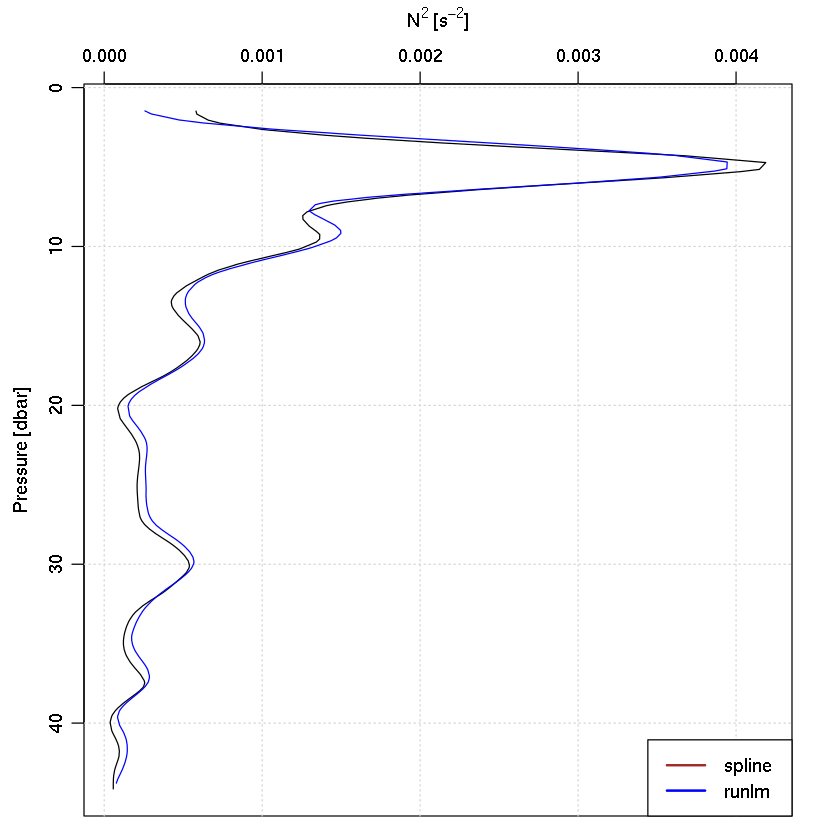
data(ctd)
rho <- swRho(ctd)
z <- swZ(ctd)
drhodz <- runlm(z, rho, deriv = 1)
g <- 9.81
rho0 <- mean(rho, na.rm = TRUE)
N2 <- -g * drhodz/rho0
plot(ctd, which = "N2")
lines(N2, -z, col = "blue")
legend("bottomright", lwd = 2, col = c("brown", "blue"), legend = c("spline",
"runlm"), bg = "white")
Example 3: T-S diagram.
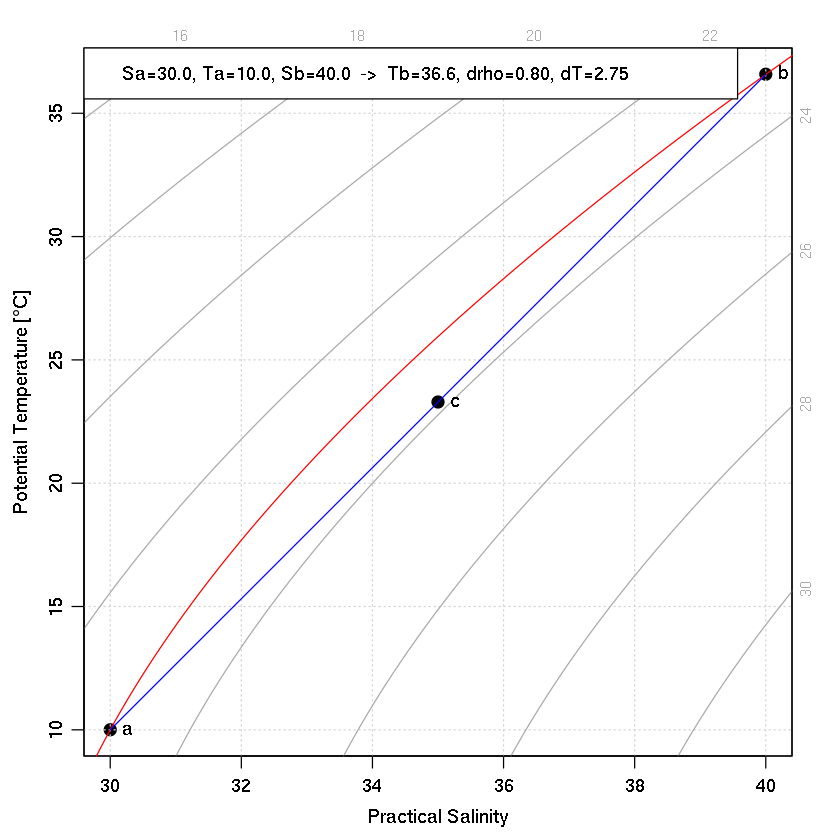
# Alter next three lines as desired; a and b are watermasses.
Sa <- 30
Ta <- 10
Sb <- 40
library(oce)
# Should not need to edit below this line
rho0 <- swRho(Sa, Ta, 0)
Tb <- uniroot(function(T) rho0-swRho(Sb,T,0), lower=0, upper=100)$root
Sc <- (Sa + Sb) /2
Tc <- (Ta + Tb) /2
## density change, and equiv temp change
drho <- swRho(Sc, Tc, 0) - rho0
dT <- drho / rho0 / swAlpha(Sc, Tc, 0)
plotTS(as.ctd(c(Sa, Sb, Sc), c(Ta, Tb, Tc), 0), pch=20, cex=2)
drawIsopycnals(levels=rho0, col="red", cex=0)
segments(Sa, Ta, Sb, Tb, col="blue")
text(Sb, Tb, "b", pos=4)
text(Sa, Ta, "a", pos=4)
text(Sc, Tc, "c", pos=4)
legend("topleft",
legend=sprintf("Sa=%.1f, Ta=%.1f, Sb=%.1f -> Tb=%.1f, drho=%.2f, dT=%.2f",
Sa, Ta, Sb, Tb, drho, dT),
bg="white")
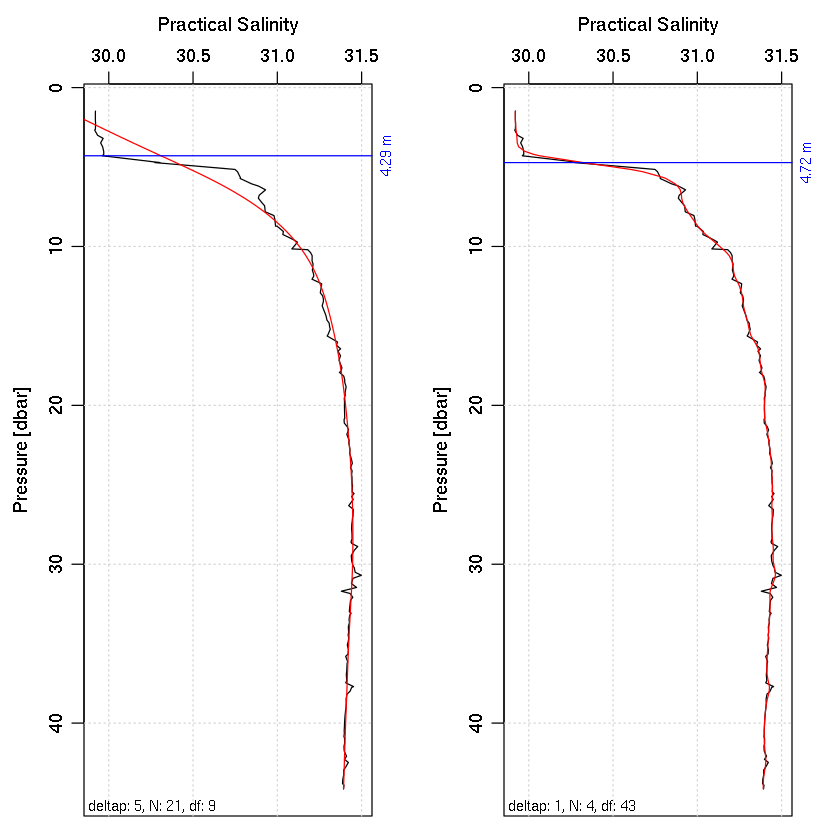
Example 4: find the halocline depth.
findHalocline <- function(ctd, deltap=5, plot=TRUE)
{
S <- ctd[['salinity']]
p <- ctd[['pressure']]
n <- length(p)
## trim df to be no larger than n/2 and no smaller than 3.
N <- deltap / median(diff(p))
df <- min(n/2, max(3, n / N))
spline <- smooth.spline(S~p, df=df)
SS <- predict(spline, p)
dSSdp <- predict(spline, p, deriv=1)
H <- p[which.max(dSSdp$y)]
if (plot) {
par(mar=c(3, 3, 1, 1), mgp=c(2, 0.7, 0))
plotProfile(ctd, xtype="salinity")
lines(SS$y, SS$x, col='red')
abline(h=H, col='blue')
mtext(sprintf("%.2f m", H), side=4, at=H, cex=3/4, col='blue')
mtext(sprintf(" deltap: %.0f, N: %.0f, df: %.0f", deltap, N, df),
side=1, line=-1, adj=0, cex=3/4)
}
return(H)
}
# Plot two panels to see influence of deltap.
par(mfrow=c(1, 2))
data(ctd)
findHalocline(ctd)
findHalocline(ctd, 1)